For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DTDMTLAGLEASDHSDLSLAKSHSWWDPTSECTIVVADPGAERELWHEYVRGAERNYRKHGVECALDMQA LRTGTDTALFCAGINDAGRVVGGLRAKGPYRNADESHAIVEWSGQPGLDAVRKMVSDRIPFGVVEMKTAW VSDDPEQSRALTDTLARMPLHSMTLLNIQFAMATAAAYVLKRWLSSGGVLATKIPATPYPDARYQTKMMW WDRSTFANHADPKQLSAYFAESQRITPRLGVADDTDSTTAAGQ*
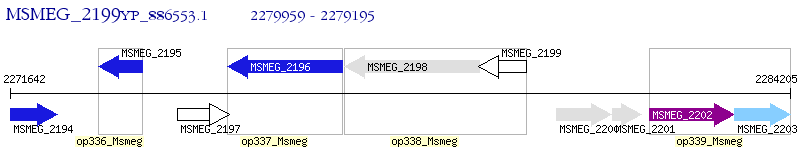
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2199 | - | - | 100% (254) | hypothetical protein MSMEG_2199 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1391c | - | 3e-71 | 50.80% (250) | hypothetical protein Mb1391c |
| M. gilvum PYR-GCK | Mflv_4375 | - | 1e-89 | 63.56% (236) | hypothetical protein Mflv_4375 |
| M. tuberculosis H37Rv | Rv1356c | - | 3e-71 | 50.80% (250) | hypothetical protein Rv1356c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3133 | - | 3e-64 | 48.95% (237) | hypothetical protein MAV_3133 |
| M. thermoresistible (build 8) | TH_0018 | - | 3e-76 | 54.07% (246) | HYPOTHETICAL PROTEIN Rv1356c |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1969 | - | 4e-78 | 56.58% (228) | hypothetical protein Mvan_1969 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2199|M.smegmatis_MC2_155 --------MDTDMTLAGLEASDHSDLSLAK-SHSWWDPTSECTIVVADPG
Mflv_4375|M.gilvum_PYR-GCK --------MDDDMTTPEFHSAAFDRASLTE-SPSYWDPATQCTVVYATPA
Mvan_1969|M.vanbaalenii_PYR-1 ------------MSITSLISADTDHFELAS-NHSWWDGDAECTLVMSQPH
TH_0018|M.thermoresistible__bu -----------MTSAAQLDGDTFGEYALPT-GISWWDPDAECTLVMSQPA
Mb1391c|M.bovis_AF2122/97 MLIAGYLTDWRIMTTAQLRPIAPQKLHFSENLSVWVSDAQ-CRLVVSQPA
Rv1356c|M.tuberculosis_H37Rv MLIAGYLTDWRIMTTAQLRPIAPQKLHFSENLSVWVSDAQ-CRLVVSQPA
MAV_3133|M.avium_104 ------------MEAAQLAAREISDGPWPETGFGWYDQATGCRVTASTPE
. : . : . * :. : *
MSMEG_2199|M.smegmatis_MC2_155 AERELWHEYVRGAERNYRKHGVECALDMQALRTGTDTALFCAGINDAGRV
Mflv_4375|M.gilvum_PYR-GCK RERELWRDYARGACASYEKHGVAAALDLDALRTGHDTVLFAALVNSAGAV
Mvan_1969|M.vanbaalenii_PYR-1 LDPDLWLEYLRGAEHSYRKHGVEAALDVDEISSGHDTAVFWAALDTTGRV
TH_0018|M.thermoresistible__bu LDRELWLDYLDGAARSYRKYGAEKALDLEAVKPGWDTSVFFAAVDRDGCV
Mb1391c|M.bovis_AF2122/97 LDPTLWNTYLQGALRAYSKHGVECTLDLDAISDGSDTQLFFAAIDIGGDV
Rv1356c|M.tuberculosis_H37Rv LDPTLWNTYLQGALRAYSKHGVECTLDLDAISDGSDTQLFFAAIDIGGDV
MAV_3133|M.avium_104 AEPELWNQYLDGALESYRKHGLDWVLDLERIRDGADTTLFFAALDPQGRV
: ** * ** * *:* .**:: : * ** :* * :: * *
MSMEG_2199|M.smegmatis_MC2_155 VGGLRAKGPYRNADESHAIVEWSGQPGLDAVRKMVSDRIPFGVVEMKTAW
Mflv_4375|M.gilvum_PYR-GCK VGGLRARGPYRSADECHAVNEWDGQDGQAAVHKMVTDRIPFGVAEMKTAW
Mvan_1969|M.vanbaalenii_PYR-1 IGGVRAKGPLTGADDSHAVVEWEGQPGLAAVRKMIDDRAPFGILEMKSAF
TH_0018|M.thermoresistible__bu IGGLRATRPLRSADESHAVVEWAGQPGESAVRKMITDRLPFGVVEMKSAW
Mb1391c|M.bovis_AF2122/97 VGGARVIGPLRSADDSHAVVEWAGNPGLSAVRKMINDRAPFGVVEVKSGW
Rv1356c|M.tuberculosis_H37Rv VGGARVIGPLRSADDSHAVVEWAGNPGLSAVRKMINDRAPFGVVEVKSGW
MAV_3133|M.avium_104 VGGTRVVGPLRSPEDSHALKEWKGNPGLPLLRYMIANRIPFGVVEVKSAW
:** *. * ..::.**: ** *: * :: *: :* ***: *:*:.:
MSMEG_2199|M.smegmatis_MC2_155 VSDDPEQSRALTDTLARMPLHSMTLLNIQFAMATAAAYVLKRWLSSGGVL
Mflv_4375|M.gilvum_PYR-GCK VTDDPDENRRLTTAIARTPLHAMTLLDIQFIVATAASYVLKRWLSSGGLL
Mvan_1969|M.vanbaalenii_PYR-1 VTDDPARNKGVTRALARSGFHAMAAMDLQFCMATSAPHVLDKWRSSGGVV
TH_0018|M.thermoresistible__bu ADGDRDRSPLLADVLARTAYPTMTVLNVQFLMATAASHVLGRWRSSGGVV
Mb1391c|M.bovis_AF2122/97 VNSDAQRSDAIAAALARALPLSMSLLGVQFVMGTAAAHALDRWRSSGGVI
Rv1356c|M.tuberculosis_H37Rv VNSDAQRSDAIAAALARALPLSMSLLGVQFVMGTAAAHALDRWRSSGGVI
MAV_3133|M.avium_104 TNSREYGNRALSTVLARTALPTMTLLGAQFVMATAAAHVLEQWQSSGGVV
. . . :: .:** :*: :. ** :.*:*.:.* :* ****::
MSMEG_2199|M.smegmatis_MC2_155 ATKIPATPYPDARYQTKMMWWDRSTFANHADPKQLSAYFAESQRITPRLG
Mflv_4375|M.gilvum_PYR-GCK ATKIPATPYPDHRYDTKLAWWDRTTFTKHAEPSQVSAFFSEQRQLAETVP
Mvan_1969|M.vanbaalenii_PYR-1 AS-IPATPYPDPRYRTKMMWWDRSTFIRHAEPAQVSKIFNEILDMQRSRV
TH_0018|M.thermoresistible__bu ASRIPATPYPDDRYRTKMMWWDRSNFINYATPEQAAKMVAEMSSLSALLD
Mb1391c|M.bovis_AF2122/97 AARIPAAAYPDERYRTKMIWWDRRTLANHAEPKQLSRMLVESRKLLRDVE
Rv1356c|M.tuberculosis_H37Rv AARIPAAAYPDERYRTKMIWWDRRTLANHAEPKQLSRMLVESRKLLRDVE
MAV_3133|M.avium_104 ASRVPAAAYPNENYRTKVMWWDRRTLAAHADPAQFKLMCAENTEILSRLS
*: :**:.**: .* **: **** .: :* * * * :
MSMEG_2199|M.smegmatis_MC2_155 VADDTDSTTAAGQ-
Mflv_4375|M.gilvum_PYR-GCK HLMGAPVTAPAL--
Mvan_1969|M.vanbaalenii_PYR-1 APEGTGSLRDNAL-
TH_0018|M.thermoresistible__bu AHRDVAAAAGTQ--
Mb1391c|M.bovis_AF2122/97 ALSATTAATAGAEQ
Rv1356c|M.tuberculosis_H37Rv ALSATTAATAGAEQ
MAV_3133|M.avium_104 A-------------