For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVLVWTGVALLGGLGAVCRFLVDRTVSARAPGGFPYGTLAVNVSGALLLGFLGGLALPANVALVVGTGFV GSYTTFSTWMLETQRLGEERQIRSAVANLVVSVVLGLAAAGLGVFLGGRL*
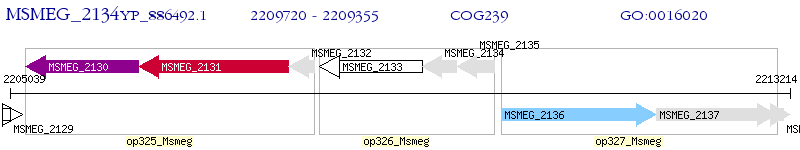
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2134 | crcB | - | 100% (121) | CrcB protein |
| M. smegmatis MC2 155 | MSMEG_2135 | - | 1e-09 | 35.92% (103) | putative CrcB protein |
| M. smegmatis MC2 155 | MSMEG_5166 | - | 4e-06 | 26.92% (130) | Na+/solute symporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3097 | ccrB | 1e-42 | 69.23% (117) | camphor resistance protein CrcB |
| M. gilvum PYR-GCK | Mflv_4408 | - | 2e-43 | 72.88% (118) | camphor resistance CrcB protein |
| M. tuberculosis H37Rv | Rv3070 | ccrB | 1e-42 | 69.23% (117) | camphor resistance protein CrcB |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2440 | - | 1e-17 | 40.16% (122) | hypothetical protein MAB_2440 |
| M. marinum M | MMAR_1589 | - | 4e-37 | 65.49% (113) | hypothetical protein MMAR_1589 |
| M. avium 104 | MAV_3977 | - | 2e-39 | 69.03% (113) | putative CrcB protein 2 |
| M. thermoresistible (build 8) | TH_2516 | - | 2e-30 | 54.55% (121) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1948 | - | 2e-40 | 70.34% (118) | CrcB protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4408|M.gilvum_PYR-GCK MTD-----ALVWVGVFLVGGLGAVCRLTVDKAVSHRARGSFPYGTLVVNI
Mvan_1948|M.vanbaalenii_PYR-1 MTA-----ALVWVGVFVVGGLGAVARLTVDKAVSQRAGWSFPLGTLAVNV
MSMEG_2134|M.smegmatis_MC2_155 MT------VLVWTGVALLGGLGAVCRFLVDRTVSARAPGGFPYGTLAVNV
Mb3097|M.bovis_AF2122/97 MTASTALTVAIWIGVMLIGGIGSVLRFLVDRSVARRLARTFPYGTLTVNI
Rv3070|M.tuberculosis_H37Rv MTASTALTVAIWIGVMLIGGIGSVLRFLVDRSVARRLARTFPYGTLTVNI
MMAR_1589|M.marinum_M MTAG---TVVVWMGVMVIGGLGSTLRFLVDRTIARRMARTFPFGTFAVNV
MAV_3977|M.avium_104 MTTA-----AVWVGVALIGGVGSVLRFVVDGAVARRVSRPFPLGTLVVNL
TH_2516|M.thermoresistible__bu MTS-----LGVWAGVAVLGGLGAVLRYLVDRTVSRRSPTRLPAGIFVVNI
MAB_2440|M.abscessus_ATCC_1997 MT---------IMIVILAGALGAVLRFVVDSAIKYRRTHRFPWTTLLINL
** * : *.:*:. * ** :: * :* : :*:
Mflv_4408|M.gilvum_PYR-GCK SGAALLGFLAGLTLPAHVA----LLAGTAFVGSYTTFSTWMLETHRLAEE
Mvan_1948|M.vanbaalenii_PYR-1 SGALLLGFLAGLALPPDLA----LLAGTAFVGSYTTFSTWMLETHRLAEE
MSMEG_2134|M.smegmatis_MC2_155 SGALLLGFLGGLALPANVA----LVVGTGFVGSYTTFSTWMLETQRLGEE
Mb3097|M.bovis_AF2122/97 TGAALLGFLAGLALPKDAA----LLAGTGFVGAYTTFSTWMLETQRLGED
Rv3070|M.tuberculosis_H37Rv TGAALLGFLAGLALPKDAA----LLAGTGFVGAYTTFSTWMLETQRLGED
MMAR_1589|M.marinum_M SGAALLGFLAGLALSRDAA----LLAGTAFVGAYTTFSTWMLETQRLSEE
MAV_3977|M.avium_104 SGAALLGFLGGLALSREAA----LLAGTAFVGAYTTFSTWMLETQRLGEE
TH_2516|M.thermoresistible__bu TGALALGLLAGAAVSPRTA----LLAGAALLGSYTTFSTWMLDTVELAER
MAB_2440|M.abscessus_ATCC_1997 TGSALIGLLAGLVIFHHAAPDLLTVLGTGFCGGYTTFSTASVETVRLIER
:*: :*:*.* .: * : *:.: *.****** ::* .* *
Mflv_4408|M.gilvum_PYR-GCK R----QIRSALGNLGLSVVLGLAAAFAGQTVGSML-
Mvan_1948|M.vanbaalenii_PYR-1 R----QIWPALGNIVISVALGLAAALLGQSLGALL-
MSMEG_2134|M.smegmatis_MC2_155 R----QIRSAVANLVVSVVLGLAAAGLGVFLGGRL-
Mb3097|M.bovis_AF2122/97 R----QMVSALANIVVSVVLGLAAALLGQWIAQI--
Rv3070|M.tuberculosis_H37Rv R----QMVSALANIVVSVVLGLAAALLGQWIAQI--
MMAR_1589|M.marinum_M R----QMWVALANIGLSVLAGLGAALLGHWIAQLL-
MAV_3977|M.avium_104 R----QLRAALANVVVSVVLGGAAAFIGQQLAQLV-
TH_2516|M.thermoresistible__bu RPRRRAVLDTVVNIVAGVVAGLGAAAVGYRVGSML-
MAB_2440|M.abscessus_ATCC_1997 R----RWTPAFYNTFGTLLGTVGACAAGLALGWVLA
* :. * : .*. * :.