For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVIDVDQLDDVANRLFGAIERGDRAAVAALWSEDVRVWHSGDPRDNDRARALRVIDWFIDATAQRHYEIL ERQFFDGGFVQQHIVHATGRDGAQIDLRVCIVIKVGADGLITRVDEYFDPEDIAPLLDQVGT*
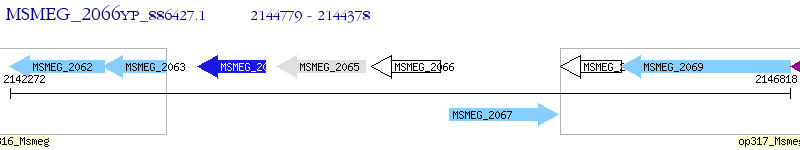
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2066 | - | - | 100% (133) | hypothetical protein MSMEG_2066 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4472 | - | 2e-40 | 64.96% (117) | hypothetical protein Mflv_4472 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1488 | - | 8e-42 | 59.09% (132) | hypothetical protein MMAR_1488 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0639 | - | 1e-45 | 65.57% (122) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2451 | - | 3e-41 | 59.09% (132) | hypothetical protein MUL_2451 |
| M. vanbaalenii PYR-1 | Mvan_1887 | - | 1e-43 | 60.15% (133) | hypothetical protein Mvan_1887 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1488|M.marinum_M --MPPPPLSS----------TEAITEVADRLFTAIENGDRAEVDRMWSPD
MUL_2451|M.ulcerans_Agy99 --MPPPPLSS----------TGAITKVADRLFTAIENGDRAEVDRMWSPD
TH_0639|M.thermoresistible__bu VTGAGHPLDGRRENTMRSDGADRITEVADRLFAAIEGGDVAAVARMWADD
MSMEG_2066|M.smegmatis_MC2_155 ----MPVID-----------VDQLDDVANRLFGAIERGDRAAVAALWSED
Mflv_4472|M.gilvum_PYR-GCK ----MPDTAV----------------VADRLFAAITRSDIDTVAQMFSPG
Mvan_1887|M.vanbaalenii_PYR-1 ----MPETDTR-----------VVGDVADRLFDAIERSDIAVVAQLFSPG
**:*** ** .* * ::: .
MMAR_1488|M.marinum_M IAVWRVGARRDNDKARALRVIDWFLSATTERRYEILDRRVFSDESASGFV
MUL_2451|M.ulcerans_Agy99 IAVWRVGARRDNDKARALRVIDWFLSATTERRYEILDRRVFSDESASGFV
TH_0639|M.thermoresistible__bu IAVWQSGDPHDRDKERALRVIDWFISGTVERRYEVLDRQVFDD----GFV
MSMEG_2066|M.smegmatis_MC2_155 VRVWHSGDPRDNDRARALRVIDWFIDATAQRHYEILERQFFDG----GFV
Mflv_4472|M.gilvum_PYR-GCK VAVWHSGDDRDCDHRRAVKVIDWFVRATVDRRYEITDRRFFDG----GFV
Mvan_1887|M.vanbaalenii_PYR-1 IAVWKSGDTRDNNHARSVKIIRWFVDATVNRRYEVLDRQMFDG----GFV
: **: * :* :: *::::* **: .*.:*:**: :*:.*.. ***
MMAR_1488|M.marinum_M QQHTLHATGRAGQVIAMRVCIVIKLDRDGVINRIDEYFDPSELAPLLD--
MUL_2451|M.ulcerans_Agy99 QQHTLHATGRAGQVIAMRVCIVIKLDRDGVINRIDEYFDPSELAPLLD--
TH_0639|M.thermoresistible__bu QQHILLARGRTGASIALRVCLVVRIGADGLINRIDEYFDPAGIAPLLAAQ
MSMEG_2066|M.smegmatis_MC2_155 QQHIVHATGRDGAQIDLRVCIVIKVGADGLITRVDEYFDPEDIAPLLDQV
Mflv_4472|M.gilvum_PYR-GCK QQHILHATARSGEAIAMRVCIVIVVGDDGLITRIDEYFDPAEMQPLLVGG
Mvan_1887|M.vanbaalenii_PYR-1 QQHVLHAATPAGATIAMRVCIVIKVGGDGLINRIDEYFDPAEIAPLLSHT
*** : * * * :***:*: :. **:*.*:****** : ***
MMAR_1488|M.marinum_M --
MUL_2451|M.ulcerans_Agy99 --
TH_0639|M.thermoresistible__bu DG
MSMEG_2066|M.smegmatis_MC2_155 GT
Mflv_4472|M.gilvum_PYR-GCK S-
Mvan_1887|M.vanbaalenii_PYR-1 SS