For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VENWRPGPVVSTDLLGPQAAGNLAATLDLDHVPGAGEVLPALWHWVYFLDWPPTAALGTDGHPRDGHFMP PIPHRRRMFAGGRLEINEPLRLGEAATRHSEVVSTAVKSGKTGELMFVTVRHEFSQGGQLRMTEEQDLVY RSDPGSSTPFTRVTEPLAERTTPWAAEPATHPALLFRFSAVTANSHRIHYDAEYTCGVEGFPALVVHGPL LVLYMAELARAHSTRPIKNFRFRLTKPVFVGDAIRVQGTPAADGAAAELAVVTGSGTVHASAQAEYA
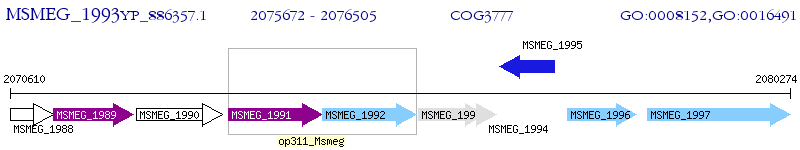
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1993 | - | - | 100% (277) | MaoC like domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3005 | - | 1e-05 | 50.00% (42) | MaoC like domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1993|M.smegmatis_MC2_155 MENWRPGPVVSTDLLGPQAAGNLAATLDLDHVPGAGEVLPALWHWVYFLD
MAV_3005|M.avium_104 --------------------------------------------------
MSMEG_1993|M.smegmatis_MC2_155 WPPTAALGTDGHPRDGHFMPPIPHRRRMFAGGRLEINEPLRLGEAATRHS
MAV_3005|M.avium_104 ------------------------------------------------MT
:
MSMEG_1993|M.smegmatis_MC2_155 EVVSTAVKSGKTGELMFVTVRHEFSQGGQLRMTEEQDLVYRSDPGSSTPF
MAV_3005|M.avium_104 ELFYDDVSVGDHIPTLTVTV------------------------------
*:. *. *. : ***
MSMEG_1993|M.smegmatis_MC2_155 TRVTEPLAERTTPWAAEPATHPALLFRFSAVTANSHRIHYDAEYTCGVEG
MAV_3005|M.avium_104 --------------------DETQMFFFSAATYNGHRIHYDKEWAR-AEG
. : :* ***.* *.****** *:: .**
MSMEG_1993|M.smegmatis_MC2_155 FPALVVHGPLLVLYMAELARAHS--TRPIKNFRFRLTKPVFVGDAIRVQG
MAV_3005|M.avium_104 YDDVLVHGPLQAALLARALGDWIGGRGRLVSFSVQNRAVAYPGEPLSFGG
: ::***** . :*. : .* .: .: *:.: . *
MSMEG_1993|M.smegmatis_MC2_155 ---------------TPAADGAAAELAVVTGSGTVHASAQAEYA
MAV_3005|M.avium_104 RVTGKRLDESGSGLVDLDIAGRRDDTVLMPGAATVQLPRREARS
* : .::.*:.**: . : :