For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSRQSLTKAHAKISELTWEPTFATPATRFGTDYTFEKAPKKDPLKQIMRSYFSMEEEKDNRVYGAMDGAI RGNMFRQVQQRWLEWQKLFLSIIPFPEISAARAMPMAIDAVPNPEIHNGLAVQMIDEVRHSTIQMNLKKL YMNNYIDPSGFDMTEKAFANNYAGTIGRQFGEGFITGDAITAANIYLTVVAETAFTNTLFVAMPDEAAAN GDYLLPTVFHSVQSDESRHISNGYSILLMALADERNRPLLERDLRYAWWNNHCVVDAAIGTFIEYGTKDR RKDRESYAEMWRRWIYDDYYRSYLLPLEKYGLTIPHDLVEEAWKRIVEKGYVHEVARFFATGWPVNYWRI DTMTDTDFEWFEHKYPGWYSKFGKWWENYNRLAYPGRNKPIAFEEVGYQYPHRCWTCMVPALIREDMIVE KVDGQWRTYCSETCYWTDAVAFRGEYEGRETPNMGRLTGFREWETLHHGKDLADIVTDLGYVRDDGKTLV GQPHLNLDPQKMWTLDDVRGNTFNSPNVLLNQMTDDERDAHVAAYRAGGVPA
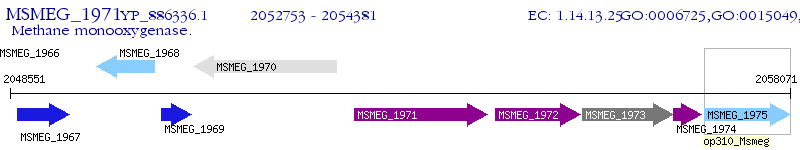
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1971 | - | - | 100% (542) | propane monooxygenase hydroxylase large subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0131 | - | 3e-91 | 38.75% (511) | monooxygenase hydroxylase, subunit A (AamH_A) |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1971|M.smegmatis_MC2_155 MSRQSLTKAHAKISELTWEPTFATPATRFGTDYTFEKAPKKDPLKQIMRS
MMAR_0131|M.marinum_M -MTASITTQHEKIKSFDWEPSYFHRDALYPTKYKIPPR-TKDPFRTLVRE
*:*. * **..: ***:: : : *.*.: .***:: ::*.
MSMEG_1971|M.smegmatis_MC2_155 YFSMEEEKDNRVYGAMDGAI-RGNMFRQVQQRWLEWQKLFLSIIPFPEIS
MMAR_0131|M.marinum_M YVGMEEEKDSRQYGALEDALSRMNNSAQVEPRFMEVLKPVLAFVDFGEYA
*..******.* ***::.*: * * **: *::* * .*::: * * :
MSMEG_1971|M.smegmatis_MC2_155 AARAMPMAIDAVPNPEIHNGLAVQMIDEVRHSTIQMNLKKLYMNNYIDPS
MMAR_0131|M.marinum_M AMKCTAMLIDTVENPELRQGYLAQMIDEVRHTNQEAYLMRYFAKHAPDPA
* :. .* **:* ***:::* .********:. : * : : :: **:
MSMEG_1971|M.smegmatis_MC2_155 GFDMTEKAFANNYAGTIGRQFGEGFITGDAITAANIYLTVVAETAFTNTL
MMAR_0131|M.marinum_M GFNSGFEMRANDPIGRPARAVFEAFMNDDPITNS-LNLQVVAETAYTNPL
**: : **: * .* . *.*:..*.** : : * ******:**.*
MSMEG_1971|M.smegmatis_MC2_155 FVAMPDEAAANGDYLLPTVFHSVQSDESRHISNGYSILLMALADERNRPL
MMAR_0131|M.marinum_M FVSLTEVAAANGDQATPSVFLSVQSDEARHMANGYATLAAVLSEPDNLPM
**::.: ****** *:** ******:**::***: * .*:: * *:
MSMEG_1971|M.smegmatis_MC2_155 LERDLRYAWWNNHCVVDAAIGTFIEYGTKDRRKDRESYAEMWRRWIYDDY
MMAR_0131|M.marinum_M LQEDFDTAFWRQHAFIDNFLGGVNDYFSKARVK---SYKEFWQQWIWEDW
*:.*: *:*.:*..:* :* . :* :* * * ** *:*::**::*:
MSMEG_1971|M.smegmatis_MC2_155 YRSYLLPLEKYGLTIPHDLVEEAWKRIVEKGYVHEVARFFATGWPVNYWR
MMAR_0131|M.marinum_M AGSYIERLEPFGLHVPR--WMDDAKRHVEWG-GHTAAMVSAALWPVHAWR
**: ** :** :*: : ** ** * * .* . *: ***: **
MSMEG_1971|M.smegmatis_MC2_155 IDTMTDTDFEWFEHKYPGWYSKFGKWWENYNRLAYPGRNKPIAFEEVGYQ
MMAR_0131|M.marinum_M SDYMTDEDFTFFEDKYPGWEKYYGDFWRAYRQMADP-RNGQLALGLFP-A
* *** ** :**.***** . :*.:*. *.::* * ** :*: .
MSMEG_1971|M.smegmatis_MC2_155 YPHRCWTCMVPALIR---EDMIVEKVD--GQWRTYCSETCYWTDAVAFRG
MMAR_0131|M.marinum_M MPPICRVCQMPAVFPRIDVNEIRLSVDKAGQRHAFCSQAC----------
* * .* :**:: : * .** ** :::**::*
MSMEG_1971|M.smegmatis_MC2_155 EYEGRETPNMGRLTGFREWETLHHGKDLADIVTDLGYVRDDGKTLVGQPH
MMAR_0131|M.marinum_M QHIFRQAP--HRHTG-KTWWEVNDGIELWRYIEINHLLRSDGKTLMGQPH
:: *::* * ** : * ::.* :* : :*.*****:****
MSMEG_1971|M.smegmatis_MC2_155 LNLDPQKMWTLDDVRGNTFNSPNVLLNQMTDDERDAHVAAYRAGGVPA
MMAR_0131|M.marinum_M VHTDERWLWTIDDIR----RTGVAIQDPLKALPAEAFTEI--------
:: * : :**:**:* .: .: : :. :*..