For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEVKIGVTDSPRELTFNSAQSPTEVEQQFTDALSSGTGVLALTDEKGRRFLVQTAKIAYVEIGAADVRRV GFGVTVESA
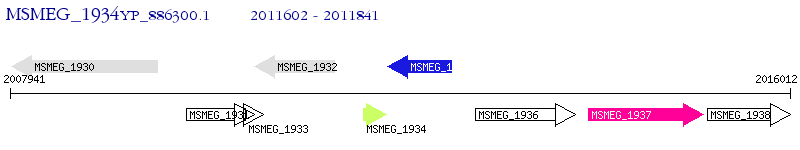
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1934 | - | - | 100% (79) | ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3234c | TB9.4 | 5e-27 | 68.35% (79) | hypothetical protein Mb3234c |
| M. gilvum PYR-GCK | Mflv_4673 | - | 8e-27 | 78.38% (74) | hypothetical protein Mflv_4673 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00814 | - | 2e-27 | 69.62% (79) | hypothetical protein MLBr_00814 |
| M. abscessus ATCC 19977 | MAB_3528c | - | 3e-21 | 63.89% (72) | hypothetical protein MAB_3528c |
| M. marinum M | MMAR_1349 | - | 4e-25 | 68.35% (79) | hypothetical protein MMAR_1349 |
| M. avium 104 | MAV_4156 | - | 7e-25 | 68.83% (77) | hypothetical protein MAV_4156 |
| M. thermoresistible (build 8) | TH_0453 | - | 3e-30 | 79.75% (79) | CONSERVED HYPOTHETICAL PROTEIN TB9.4 |
| M. ulcerans Agy99 | MUL_2530 | - | 7e-25 | 67.95% (78) | hypothetical protein MUL_2530 |
| M. vanbaalenii PYR-1 | Mvan_1794 | - | 2e-28 | 78.48% (79) | hypothetical protein Mvan_1794 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4673|M.gilvum_PYR-GCK MEVKIGVTDSPRELVFASAQTPAEVEKLVADALSGEPGVLGLTDEKGRRF
Mvan_1794|M.vanbaalenii_PYR-1 MEVKIGVTDSPRELVFASAQTPAEVEKLVTDALSGDPGVLGLTDEKGRRF
TH_0453|M.thermoresistible__bu VEVKIGVSDSPRELVFNSAQSPSEVEKLVTDALSKDSGVLSLTDDKGRRF
MSMEG_1934|M.smegmatis_MC2_155 MEVKIGVTDSPRELTFNSAQSPTEVEQQFTDALSSGTGVLALTDEKGRRF
MMAR_1349|M.marinum_M MEVKIGITDSPRELVFGSAQTPGEVEELVSAALRDDSGLLSLDDERGRRF
MUL_2530|M.ulcerans_Agy99 MEVKIGITDSPRELVFGSAQTPGEVEELVSAALRDDSGLLSLDDERGRRF
Mb3234c|M.bovis_AF2122/97 MEVKIGITDSPRELVFSSAQTPSEVEELVSNALRDDSGLLTLTDERGRRF
MLBr_00814|M.leprae_Br4923 MEVKIGITDSPRELTFSSAQTPGEIEELVSAALREGLGLLVLTDERGRRF
MAV_4156|M.avium_104 MEVKIGITDSPRELVLTSTQTPDEVEELVSAALSDGSDLLRLSDERGRRF
MAB_3528c|M.abscessus_ATCC_199 MEIKIGVTDSPRELVISSDQAPADVEKVVTAALSKESDVLNLTDQKGRKY
:*:***::******.: * *:* ::*: .: ** .:* * *::**::
Mflv_4673|M.gilvum_PYR-GCK LVQSTRIAYVEIGAADSRRVGFGVGAVGSEAV------------------
Mvan_1794|M.vanbaalenii_PYR-1 LVQSARIAYVEIGAADSRRVGFGVGVVGAEATK-----------------
TH_0453|M.thermoresistible__bu LVQTSRIAYVEIGAADVRRVGFGVGVVGAEAVKGGSGKSGSKSGTAKSGA
MSMEG_1934|M.smegmatis_MC2_155 LVQTAKIAYVEIGAADVRRVGFGVTVESA---------------------
MMAR_1349|M.marinum_M LVAAAKIAYVEIGVADARRVGFGVGAEA--------ARS-----------
MUL_2530|M.ulcerans_Agy99 LVAAAKIAYVEIGVADARRVGFGVGAEA--------VRS-----------
Mb3234c|M.bovis_AF2122/97 LIHTARIAYVEIGVADARRVGFGVGVDAAAGSAGKVATSG----------
MLBr_00814|M.leprae_Br4923 LIHGAKIAYVEIGVADARRVGFGIGAES--------ATNG----------
MAV_4156|M.avium_104 LVHTSKIAYVEIGIADVRRVGFGVGAEVVG----KHAGKGR---------
MAB_3528c|M.abscessus_ATCC_199 LIAAARIAYVEIGVADARRVGFSS--------------------------
*: ::******* ** *****.
Mflv_4673|M.gilvum_PYR-GCK ----
Mvan_1794|M.vanbaalenii_PYR-1 ----
TH_0453|M.thermoresistible__bu VKNA
MSMEG_1934|M.smegmatis_MC2_155 ----
MMAR_1349|M.marinum_M ----
MUL_2530|M.ulcerans_Agy99 ----
Mb3234c|M.bovis_AF2122/97 ----
MLBr_00814|M.leprae_Br4923 ----
MAV_4156|M.avium_104 ----
MAB_3528c|M.abscessus_ATCC_199 ----