For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRVYIPTTLAALQRLVTDRSLLVTNGTAFAVTPTLREAYAEGDDDELAEVALRDAALASMRLLATGGESD LPPRRAVVVADVPGAQPRPDLDDAVVRLPGPVPIDSVIAVYVDNADAEPAVRAAVDVIDEADLGDGDAEL TVGDAQDHDLAWYASQELPFLLELL
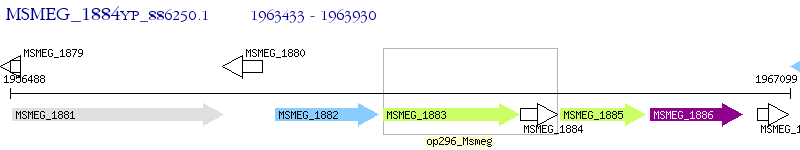
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1884 | - | - | 100% (165) | hypothetical protein MSMEG_1884 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3260c | - | 2e-63 | 74.85% (167) | hypothetical protein Mb3260c |
| M. gilvum PYR-GCK | Mflv_4700 | - | 4e-59 | 69.82% (169) | hypothetical protein Mflv_4700 |
| M. tuberculosis H37Rv | Rv3231c | - | 2e-63 | 74.85% (167) | hypothetical protein Rv3231c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3550c | - | 1e-57 | 67.47% (166) | hypothetical protein MAB_3550c |
| M. marinum M | MMAR_1313 | - | 5e-62 | 70.91% (165) | hypothetical protein MMAR_1313 |
| M. avium 104 | MAV_4194 | - | 2e-62 | 74.40% (168) | hypothetical protein MAV_4194 |
| M. thermoresistible (build 8) | TH_0651 | - | 1e-63 | 72.73% (165) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2567 | - | 9e-62 | 70.30% (165) | hypothetical protein MUL_2567 |
| M. vanbaalenii PYR-1 | Mvan_1766 | - | 8e-59 | 70.83% (168) | hypothetical protein Mvan_1766 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3260c|M.bovis_AF2122/97 ------------MTQVYIPATLAMLQRLVADGALWPVNGTAFAVTPTLRE
Rv3231c|M.tuberculosis_H37Rv ------------MTQVYIPATLAMLQRLVADGALWPVNGTAFAVTPTLRE
MMAR_1313|M.marinum_M ------------MIQVYVPATLAMLQQLVADGSLWPVNGTVFAVTPTLRE
MUL_2567|M.ulcerans_Agy99 ------------MIQVYVPATLAMLQQLVADGSLWPVNGTVFAVTPTLRE
MAV_4194|M.avium_104 ------------MATVYIPATLAMLQRLVADGSLAAVNGTAFALTPALRE
Mflv_4700|M.gilvum_PYR-GCK MTGSSAAGGYPLVVRVYVPATLAMLQRLVADGSLRPVNGTAFAVTPTLRE
Mvan_1766|M.vanbaalenii_PYR-1 -------------MRVFVPATLAMLQRLVAAESFRPVNGTAFAVTPALRE
TH_0651|M.thermoresistible__bu -------------VRIFVPATVAMLQQLVADGSLSAVNGTAFAVTPALRE
MSMEG_1884|M.smegmatis_MC2_155 -------------MRVYIPTTLAALQRLVTDRSLLVTNGTAFAVTPTLRE
MAB_3550c|M.abscessus_ATCC_199 -----------MPMQVYIPVTLSMLRGLVADGELWPVNGTAFAVTPALRE
:::*.*:: *: **: : .***.**:**:***
Mb3260c|M.bovis_AF2122/97 SYAEGDDEELAEVALREAALASLRLLA-ADIGATADALPPRRAVLAAEVD
Rv3231c|M.tuberculosis_H37Rv SYAEGDDEELAEVALREAALASLRLLA-ADIGATADALPPRRAVLAAEVD
MMAR_1313|M.marinum_M AYAQGDDDELAEVALQEAALASLRLLA-NDE---DEKLPPRRAVLTAEVD
MUL_2567|M.ulcerans_Agy99 AYAQGDDDELAEVALQEAALASLRLLA-NDE---DEKLPPRRAVLTAEVD
MAV_4194|M.avium_104 AYAEGDDDELAEVALREAALASLRLLAGAEHEAEHGALPPRRAVLAADVD
Mflv_4700|M.gilvum_PYR-GCK SYSEGDDEELGEVARREAALASLRLLG-HED---TGELPPRRVVVETDID
Mvan_1766|M.vanbaalenii_PYR-1 AYAEGDEDELADVALREAALASLRLLD-DEP---AEGLPPRRVVVELDLD
TH_0651|M.thermoresistible__bu AYAEGDDEELAHVALREAALASLRLLS-AEE---GSGAPMRRAVVEAEID
MSMEG_1884|M.smegmatis_MC2_155 AYAEGDDDELAEVALRDAALASMRLLATGGE----SDLPPRRAVVVADVP
MAB_3550c|M.abscessus_ATCC_199 AYSAGDEDELSEVAMREAALASLRLLATEDG---DDGLPARRAVLVADAE
:*: **::**..** ::*****:*** * **.*: :
Mb3260c|M.bovis_AF2122/97 D----ATYRPDLDDAVVRLAGPITIDQVVAAYVDNAGAEPAVMAAIAVID
Rv3231c|M.tuberculosis_H37Rv D----ATYRPDLDDAVVRLAGPITIDQVVAAYVDNAGAEPAVMAAIAVID
MMAR_1313|M.marinum_M D----AIYRPDLDDAVVRLSAPIAMDQVVAAYVDNVAAEPAVSAAIAVID
MUL_2567|M.ulcerans_Agy99 D----AIYRPDLDDAVVRLSAPIAMDQVVAAYVDNVAAEPAVSAAIAVID
MAV_4194|M.avium_104 D----LTYRPDLDDAVVRLAGPVRIEDVIAAYVDNAAAEPAVAKAIEAVD
Mflv_4700|M.gilvum_PYR-GCK EAADGLTLRPDLDDAVVRLSGEIDLGRVIAAYVDNQAAEAAVRAAVEVID
Mvan_1766|M.vanbaalenii_PYR-1 D-ADGITFRPDLDDAVVRIAGDVPIGRVVAAYVDNAAAEPAVLAAVEVID
TH_0651|M.thermoresistible__bu G----ATPRPDLDDAVVRLSGPIRLDDVIAAYVDNSDAETAVREAVEVID
MSMEG_1884|M.smegmatis_MC2_155 G----AQPRPDLDDAVVRLPGPVPIDSVIAVYVDNADAEPAVRAAVDVID
MAB_3550c|M.abscessus_ATCC_199 E----AKLRPDLDDAVVRLPGAIPMSAVVAAHIDLAQAEPDVVRAAGVID
**********:.. : : *:*.::* **. * * .:*
Mb3260c|M.bovis_AF2122/97 AADLGDEDAELVVGDAQDHDLAWYANQELPFLLDLL
Rv3231c|M.tuberculosis_H37Rv AADLGDEDAELVVGDAQDHDLAWYANQELPFLLDLL
MMAR_1313|M.marinum_M AADLGDEDADLVVGDAQDHDLAWYANQELPFLLDLL
MUL_2567|M.ulcerans_Agy99 AAELGDEDADLVVGDAQDHDLAWYANQELPFLLDLL
MAV_4194|M.avium_104 AADLGDEDADLIVGDAQDHDLAWYASQELPFLLELL
Mflv_4700|M.gilvum_PYR-GCK AADLGNEDAELTVGDAQDHDLAWYAPQELPFLLDLL
Mvan_1766|M.vanbaalenii_PYR-1 SADLGDEDAELTVGDAQDHDLAWYAPQELNFLLDLL
TH_0651|M.thermoresistible__bu AADLGDEDAELLVGDAQDHDLAWYATQELPFLLDLL
MSMEG_1884|M.smegmatis_MC2_155 EADLGDGDAELTVGDAQDHDLAWYASQELPFLLELL
MAB_3550c|M.abscessus_ATCC_199 AADLGDEDAELVVGDAQDHDLAWYATQELPFLLDLL
*:**: **:* ************* *** ***:**