For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLSKLLRLGEGRMVKRLRKVADYVNALSDDVEKLSDAELRAKTEEFKKRVADGEDLDDLLPEAFAVAREA AWRVLNQRHFDVQVMGGAALHFGNVAEMKTGEGKTLTAVLPSYLNALSGKGVHVVTVNDYLARRDSEWMG RVHRFLGLDVGVILSGMTPDERRAAYAADITYGTNNEFGFDYLRDNMAHSVDDMVQRGHNFAIVDEVDSI LIDEARTPLIISGPADGASHWYQEFARIVPMMEKDVHYEVDLRKRTVGVHELGVEFVEDQLGIDNLYEAA NSPLVSYLNNALKAKELFQRDKDYIVRNGEVLIVDEFTGRVLMGRRYNEGMHQAIEAKERVEIKAENQTL ATITLQNYFRLYDKLSGMTGTAETEAAELHEIYKLGVVPIPTNKPMVRQDQSDLIYKTEEAKFLAVVDDV AERHAKGQPVLIGTTSVERSEYLSKMLTKRRVPHNVLNAKYHEQEANIIAEAGRRGAVTVATNMAGRGTD IVLGGNVDFLADKRLRERGLDPVETPEEYEAAWHEVLPQVKAECAKEAEQVIEAGGLYVLGTERHESRRI DNQLRGRSGRQGDPGESRFYLSLGDELMRRFNGATLETLLTRLNLPDDVPIEAKMVSRAIKSAQTQVEQQ NFEVRKNVLKYDEVMNQQRKVIYAERRRILEGENLAEQAHKMLVDVITAYVDGATAEGYAEDWDLETLWT ALKTLYPVGIDHRDLIDSDAVGEPGELTREELLDALIKDAERAYAEREKQIEAIAGEGAMRQLERNVLLN VIDRKWREHLYEMDYLKEGIGLRAMAQRDPLVEYQREGYDMFVGMLEALKEESVGFLFNVQVEAAPQQPQ VAPQAPPPTLSEFAAAAAAKASDSAAKPDSGSVATKERAEAERPAPALRAKGIDNEAPPLTYTGPSEDGT AQVQRSGNGGRHAAPAGGSRRERREAARKQAKADRPAKSHRKG
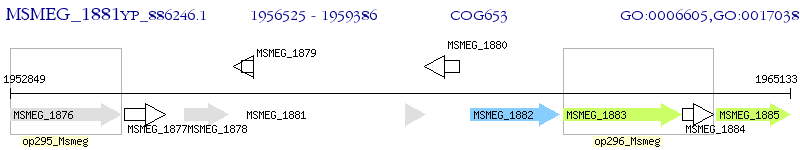
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1881 | secA | - | 100% (953) | preprotein translocase subunit SecA |
| M. smegmatis MC2 155 | MSMEG_3654 | - | e-121 | 40.33% (667) | preprotein translocase subunit SecA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3268c | secA1 | 0.0 | 83.21% (959) | preprotein translocase subunit SecA |
| M. gilvum PYR-GCK | Mflv_4705 | - | 0.0 | 79.98% (949) | preprotein translocase subunit SecA |
| M. tuberculosis H37Rv | Rv3240c | secA1 | 0.0 | 83.21% (959) | preprotein translocase subunit SecA |
| M. leprae Br4923 | MLBr_00779 | secA | 0.0 | 78.29% (958) | preprotein translocase subunit SecA |
| M. abscessus ATCC 19977 | MAB_3580c | - | 0.0 | 78.59% (953) | preprotein translocase subunit SecA |
| M. marinum M | MMAR_1305 | secA1 | 0.0 | 82.01% (956) | preprotein translocase SecA1 1 subunit |
| M. avium 104 | MAV_2894 | - | 1e-118 | 39.51% (653) | preprotein translocase subunit SecA |
| M. thermoresistible (build 8) | TH_0994 | secA | 0.0 | 90.58% (807) | PROBABLE PREPROTEIN TRANSLOCASE SECA1 1 SUBUNIT |
| M. ulcerans Agy99 | MUL_2576 | secA1 | 0.0 | 81.28% (956) | preprotein translocase subunit SecA |
| M. vanbaalenii PYR-1 | Mvan_1762 | - | 0.0 | 80.51% (949) | preprotein translocase subunit SecA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4705|M.gilvum_PYR-GCK MLSKLLRIGEGRMVKRLKGVSDYVN--TLSDDMEKLS-----DADLRAKT
Mvan_1762|M.vanbaalenii_PYR-1 MLSKLLRLGEGRMVKRLKGVADYVN--TLSDDIEKLS-----DAELRGKT
MSMEG_1881|M.smegmatis_MC2_155 MLSKLLRLGEGRMVKRLRKVADYVN--ALSDDVEKLS-----DAELRAKT
Mb3268c|M.bovis_AF2122/97 MLSKLLRLGEGRMVKRLKKVADYVG--TLSDDVEKLT-----DAELRAKT
Rv3240c|M.tuberculosis_H37Rv MLSKLLRLGEGRMVKRLKKVADYVG--TLSDDVEKLT-----DAELRAKT
MLBr_00779|M.leprae_Br4923 MLSKLLRLGEGRIVKRLKRVAEYVN--TLSDNVEKLT-----DVELKAKT
MMAR_1305|M.marinum_M MLSKLLRVGEGRMVKRLKKVADYVE--SLSGDVEKLT-----DAELRAKT
MUL_2576|M.ulcerans_Agy99 MLSKLLRVGEGRMVKRLKKVADYVE--SLSGDVEKLT-----DAELRAKT
MAB_3580c|M.abscessus_ATCC_199 MLSKLLRLGEGRMVKRLKGVADYVN--TLSDDIEKLS-----DAELQAKT
TH_0994|M.thermoresistible__bu VLSKLLRLGEGRMVKRLRKVADYVN--TLSDDVEKLS-----DAELRAKT
MAV_2894|M.avium_104 -MPKTNRAQPGRLSSRFWRLLGASTEKNRSRSLTLVTDSSEYDDEAAGLT
:.* * **: .*: : * .: :: * : . *
Mflv_4705|M.gilvum_PYR-GCK DEFRRRLADGK--EDLDDL--MPEAFAVAREAAWRVLNQKHFDVQVMGGA
Mvan_1762|M.vanbaalenii_PYR-1 DEFRARLAGGK--EDLDDV--MPEAFAVVREAAWRVLNQRHFDVQIMGGA
MSMEG_1881|M.smegmatis_MC2_155 EEFKKRVADG---EDLDDL--LPEAFAVAREAAWRVLNQRHFDVQVMGGA
Mb3268c|M.bovis_AF2122/97 DEFKRRLADQKNPETLDDL--LPEAFAVAREAAWRVLDQRPFDVQVMGAA
Rv3240c|M.tuberculosis_H37Rv DEFKRRLADQKNPETLDDL--LPEAFAVAREAAWRVLDQRPFDVQVMGAA
MLBr_00779|M.leprae_Br4923 DEFKQRLADEKNPESLDDL--LPEAFAVAREAAWRVLDQRPFDVQVMGGA
MMAR_1305|M.marinum_M DEFKKRHAEG---ESLDEL--LPEAFAVAREAAWRVLDQRPFEVQLMGAA
MUL_2576|M.ulcerans_Agy99 DEFKKRHAEG---ESLDEL--LPEAFAVAREAAWRVLGQRPFEVQLMGAA
MAB_3580c|M.abscessus_ATCC_199 GEFKGRLEKG---ETLDDL--MPEAFAVAREASWRVLSQRHFDVQVMGGA
TH_0994|M.thermoresistible__bu DEFKKRVAAG---EDLDDL--LPEAFAVAREAAWRVLSQRHFDVQVMGGA
MAV_2894|M.avium_104 DEQLRKAAGLLNLEDLAESEDIPQFLAIAREAAERATGLRPFDVQLLGAL
* : * * : :*: :*:.***: *. . : *:**::*.
Mflv_4705|M.gilvum_PYR-GCK ALHFGNVAEMKTGEGKTLTSVLPAYLNALTGKGVHIVTVNEYLAKRDAEQ
Mvan_1762|M.vanbaalenii_PYR-1 ALHFGNVAEMKTGEGKTLTSVLPAYLNALPGKGVHIVTVNEYLAKRDAEQ
MSMEG_1881|M.smegmatis_MC2_155 ALHFGNVAEMKTGEGKTLTAVLPSYLNALSGKGVHVVTVNDYLARRDSEW
Mb3268c|M.bovis_AF2122/97 ALHLGNVAEMKTGEGKTLTCVLPAYLNALAGNGVHIVTVNDYLAKRDSEW
Rv3240c|M.tuberculosis_H37Rv ALHLGNVAEMKTGEGKTLTCVLPAYLNALAGNGVHIVTVNDYLAKRDSEW
MLBr_00779|M.leprae_Br4923 ALHLGNVAEMKTGEGKTLTCVLPAYLNALAGKGVHIVTVNDYLAKRDSEW
MMAR_1305|M.marinum_M ALHLGNVAEMKTGEGKTLTSVLPAYLNGIGGKGVHIVTVNDYLAKRDSEW
MUL_2576|M.ulcerans_Agy99 ALHLGNVAEMKTGEGKTLTSVLPAYLNGIGGKGVHIVTVNDYLAKRDSEW
MAB_3580c|M.abscessus_ATCC_199 ALHAGNIAEMKTGEGKTLTCVLPAYLNALAGKGTHVVTVNDYLAKRDSEW
TH_0994|M.thermoresistible__bu ALHFGNVAEMKTGEGKTLTAVLPAYLNALTGKGVHIVTVNDYLAKRDSEW
MAV_2894|M.avium_104 RMLAGDVIEMATGEGKTLAGAIAAAGYALAGRHVHVVTINDYLARRDAEW
: *:: ** *******: .:.: .: *. .*:**:*:***:**:*
Mflv_4705|M.gilvum_PYR-GCK MGRVHRFLGLDVDVILGTLTPDQRRAAYNADITYGTNWELGFDYLRDNMA
Mvan_1762|M.vanbaalenii_PYR-1 MGRVHRFLGLDVDVILGTLTPDQRRAAYNADITYGTNWELGFDYLRDNMA
MSMEG_1881|M.smegmatis_MC2_155 MGRVHRFLGLDVGVILSGMTPDERRAAYAADITYGTNNEFGFDYLRDNMA
Mb3268c|M.bovis_AF2122/97 MGRVHRFLGLQVGVILATMTPDERRVAYNADITYGTNNEFGFDYLRDNMA
Rv3240c|M.tuberculosis_H37Rv MGRVHRFLGLQVGVILATMTPDERRVAYNADITYGTNNEFGFDYLRDNMA
MLBr_00779|M.leprae_Br4923 MGRVHRFLGLQVGVILASMTPDERRVAYNADITYGTNNEFGFDYLRDNMA
MMAR_1305|M.marinum_M MGRVHRFLGLDVGVILAQMTPEERRVAYNADITYGTNNEFGFDYLRDNMA
MUL_2576|M.ulcerans_Agy99 MGRVHRFLGLDVGVILAQMTPEERRVAYNADITYGTNNEFGFDYLRDNMA
MAB_3580c|M.abscessus_ATCC_199 MGRVHRFLGLDVGVILSQMTPLERRDAYNADITYGTNNEFGFDYLRDNMT
TH_0994|M.thermoresistible__bu MGRVHRFLGLEVGVILSGMTPDQRRAAYNADITYGTNNEFGFDYLRDNMA
MAV_2894|M.avium_104 MGPLIEAMGLTVGWITAESSSEERRAAYGCDVTYASVNEIGFDVLRDQLV
** : . :** *. * . :. :** ** .*:**.: *:*** ***::.
Mflv_4705|M.gilvum_PYR-GCK LRLEDCVQRGHNFAIVDEVDSILIDEARTPLIISGPAD-GGSNWYTEFAR
Mvan_1762|M.vanbaalenii_PYR-1 LRLEDCVQRGHHFAIVDEVDSILIDEARTPLIISGPAD-GGSNWYTEFAR
MSMEG_1881|M.smegmatis_MC2_155 HSVDDMVQRGHNFAIVDEVDSILIDEARTPLIISGPAD-GASHWYQEFAR
Mb3268c|M.bovis_AF2122/97 HSLDDLVQRGHHYAIVDEVDSILIDEARTPLIISGPAD-GASNWYTEFAR
Rv3240c|M.tuberculosis_H37Rv HSLDDLVQRGHHYAIVDEVDSILIDEARTPLIISGPAD-GASNWYTEFAR
MLBr_00779|M.leprae_Br4923 HSLDDLVQRGHSYAIVDEVDSILIDEARTPLIISGSAD-GASNWYTEFAR
MMAR_1305|M.marinum_M HTLDDCVQRGHNFAIVDEVDSILIDEARTPLIISGPAD-GSSNWYTEFAR
MUL_2576|M.ulcerans_Agy99 HTLDDCVQRGHNFVIVDEVDSILIDEARTPLIISGPAD-GSSNWYTEFAR
MAB_3580c|M.abscessus_ATCC_199 HSLDELVQRGHAFAIVDEVDSILIDEARTPLIISGPAD-GASNWYTEFAR
TH_0994|M.thermoresistible__bu HSLADLVQRGHHYAIVDEVDSILIDEARTPLIISGPADSGATNWYVEFAR
MAV_2894|M.avium_104 TDVADLVSPNPDVALIDEADSVLVDEALVPLVLAGTTH--RETPRLEIIK
: : *. . .::**.**:*:*** .**:::*.:. *: :
Mflv_4705|M.gilvum_PYR-GCK LAPLMEKDVHYEVDIKKRVVGINEIGVEFVEDQLG-IENLYEAANSPLIS
Mvan_1762|M.vanbaalenii_PYR-1 LAPLMKPDVHYEVDIKKRVVGINEAGVEFVEDQLG-IENLYEAANSPLIS
MSMEG_1881|M.smegmatis_MC2_155 IVPMMEKDVHYEVDLRKRTVGVHELGVEFVEDQLG-IDNLYEAANSPLVS
Mb3268c|M.bovis_AF2122/97 LAPLMEKDVHYEVDLRKRTVGVHEKGVEFVEDQLG-IDNLYEAANSPLVS
Rv3240c|M.tuberculosis_H37Rv LAPLMEKDVHYEVDLRKRTVGVHEKGVEFVEDQLG-IDNLYEAANSPLVS
MLBr_00779|M.leprae_Br4923 LVRLMDKDVHYEVDLRKRTVGVHEKGMEFVEDQLG-IDNLYEVANSPLVS
MMAR_1305|M.marinum_M LAPLMEKDTHYEVDLRKRTVGVHELGVEFVEDQLG-IDNLYEAANSPLVS
MUL_2576|M.ulcerans_Agy99 LAPLMEKDTHYEVDLRKRTVGVHELGVEFVEDQLG-IDNLYEAANSPLVS
MAB_3580c|M.abscessus_ATCC_199 IVPLMEKDTHYEVDIRKRTIGVHELGVEFVEDQLG-IDNLYEAANSPLVS
TH_0994|M.thermoresistible__bu IAPLMQKDVHYEVDLRKRTVGVHELGVEFVEDQLG-IDNLYEAANSPLVS
MAV_2894|M.avium_104 LVGELEAGTDYDTDADSRNVHLTDVGARKVEKALGGIDLYSEEHVGTTLT
:. :. ...*:.* .* : : : * . **. ** *: * .. ::
Mflv_4705|M.gilvum_PYR-GCK YLNNAIKAKELFERDKHYIVRNGEVFIVDEFTGRMLVGRRYNEGLHQAIE
Mvan_1762|M.vanbaalenii_PYR-1 YLNNAIKAKELFERDKHYIVRNGEVFIVDEFTGRMLVGRRYNEGLHQAIE
MSMEG_1881|M.smegmatis_MC2_155 YLNNALKAKELFQRDKDYIVRNGEVLIVDEFTGRVLMGRRYNEGMHQAIE
Mb3268c|M.bovis_AF2122/97 YLNNALKAKELFSRDKDYIVRDGEVLIVDEFTGRVLIGRRYNEGMHQAIE
Rv3240c|M.tuberculosis_H37Rv YLNNALKAKELFSRDKDYIVRDGEVLIVDEFTGRVLIGRRYNEGMHQAIE
MLBr_00779|M.leprae_Br4923 YLNNALKAKELFNRDKDYIVRDGEVLIVDEFTGRVLIGRRYNEGMHQAIE
MMAR_1305|M.marinum_M YLNNALKAKELFNRDKDYIVRNGEVLIVDEFTGRVLIGRRYNEGMHQAIE
MUL_2576|M.ulcerans_Agy99 YLNNALKAKELFNRDKDYIVRNGEVLIVDEFTGRVLIGRRYSEGMHQAIE
MAB_3580c|M.abscessus_ATCC_199 YLNNAIKAKELFTRDKDYIVREGEVLIVDEFTGRVLMGRRYNEGMHQAIE
TH_0994|M.thermoresistible__bu YLNNALKAKELFHRDKDYIVRNGEVLIVDEFTGRVLVGRRYNEGMHQAIE
MAV_2894|M.avium_104 EVNVALHAHVLLQRDVHYIVRDDAVHLINASRGRIAQLQRWPDGLQAAVE
:* *::*: *: ** .****:. * ::: **: :*: :*:: *:*
Mflv_4705|M.gilvum_PYR-GCK AKEHVEIKAENQTVAQITLQNYFRMYNKLAGMTGTAETEAAELHEIYKLG
Mvan_1762|M.vanbaalenii_PYR-1 AKEHVEIKAENQTVAQVTLQNYFRMYEKLAGMTGTAETEAAELHEIYKLG
MSMEG_1881|M.smegmatis_MC2_155 AKERVEIKAENQTLATITLQNYFRLYDKLSGMTGTAETEAAELHEIYKLG
Mb3268c|M.bovis_AF2122/97 AKEHVEIKAENQTLATITLQNYFRLYDKLAGMTGTAQTEAAELHEIYKLG
Rv3240c|M.tuberculosis_H37Rv AKEHVEIKAENQTLATITLQNYFRLYDKLAGMTGTAQTEAAELHEIYKLG
MLBr_00779|M.leprae_Br4923 AKEHVEIKAENQTLATITLQNYFRLYDKLAGMTGTAQTEAAELHEIYKLG
MMAR_1305|M.marinum_M AKEHVEIKAENQTLATITLQNYFRLYNKLAGMTGTAQTEAAELHEIYKLG
MUL_2576|M.ulcerans_Agy99 AKEHVEIKAENQTLATITLQNYFRLYNKHAGMTGTAQTEAAELHEIYKLG
MAB_3580c|M.abscessus_ATCC_199 AKERVEIKAENQTLATITLQNYFRLYDKLAGMTGTAQTEAAELNEIYKLG
TH_0994|M.thermoresistible__bu AKEGVEIKAENQTLATITLQNYFRLYDKLAGMTGTAETEAAELHEIYKLG
MAV_2894|M.avium_104 AKEGIETTETGEVLDTITVQALINRYATVCGMTGTALAAGEQLRQFYKLG
*** :* . .:.: :*:* :. * . .****** : . :*.::****
Mflv_4705|M.gilvum_PYR-GCK VVPIPTNRPMIRADQSDLIYKTEEAKYIAVVDDVVERYEKGQPVLIGTTS
Mvan_1762|M.vanbaalenii_PYR-1 VVPIPTNRPMVRKDQSDLIYKTEEAKYIAVVDDVAERYEKGQPVLIGTTS
MSMEG_1881|M.smegmatis_MC2_155 VVPIPTNKPMVRQDQSDLIYKTEEAKFLAVVDDVAERHAKGQPVLIGTTS
Mb3268c|M.bovis_AF2122/97 VVSIPTNMPMIREDQSDLIYKTEEAKYIAVVDDVAERYAKGQPVLIGTTS
Rv3240c|M.tuberculosis_H37Rv VVSIPTNMPMIREDQSDLIYKTEEAKYIAVVDDVAERYAKGQPVLIGTTS
MLBr_00779|M.leprae_Br4923 VVAIPTNKPMIREDRSDLIYKTEEAKYMAVVDDVAERYEKGQPVLIGTTS
MMAR_1305|M.marinum_M VVPIPTNRPMVREDQSDLIYKTEEAKYIAVVDDVAERYEKGQPVLIGTTS
MUL_2576|M.ulcerans_Agy99 VVPIPTNRPMVREDQSDLIYKTEEAKYIAVVDDVAERYEKGQPVLIGTTS
MAB_3580c|M.abscessus_ATCC_199 VVSIPTNRPMVRKDQSDLIYKTEEAKYIAVVDDVVERYEKGQPVLIGTTS
TH_0994|M.thermoresistible__bu VVPIPTNKPMIREDRADLIYKTEEAKFLAVADDIAERYQKGQPVLVGTTS
MAV_2894|M.avium_104 VSPIPPNKPNIREDEADRVYITAAAKNDAIVEHIIEVHETGQPVLVGTRD
* .**.* * :* *.:* :* * ** *:.:.: * : .*****:** .
Mflv_4705|M.gilvum_PYR-GCK VERSEFLSRQFEKRRIPHNVLNAKFHEQEAGIIAEAGRLGAITVATNMAG
Mvan_1762|M.vanbaalenii_PYR-1 VERSEFLSRQFEKRRIPHNVLNAKYHEQEAGIVAEAGRLGAITVATNMAG
MSMEG_1881|M.smegmatis_MC2_155 VERSEYLSKMLTKRRVPHNVLNAKYHEQEANIIAEAGRRGAVTVATNMAG
Mb3268c|M.bovis_AF2122/97 VERSEYLSRQFTKRRIPHNVLNAKYHEQEATIIAVAGRRGGVTVATNMAG
Rv3240c|M.tuberculosis_H37Rv VERSEYLSRQFTKRRIPHNVLNAKYHEQEATIIAVAGRRGGVTVATNMAG
MLBr_00779|M.leprae_Br4923 VERSEYLSRQFTKRHIPHNVLNAKYHEQEAGIVAVAGRRGGVTVATNMAG
MMAR_1305|M.marinum_M VERSEYLSRQFTKRRIPHNVLNAKYHEQEAGIIAEAGRRGAITVATNMAG
MUL_2576|M.ulcerans_Agy99 VERSEYLSRQFTKRRIPHNVLNAKYHEQEAGIIAEAGRRGAITVATNMAG
MAB_3580c|M.abscessus_ATCC_199 VERSEYLSRQFTKRRVPHNVLNAKYHEQEAAIIAEAGRRGAITVATNMAG
TH_0994|M.thermoresistible__bu VERSEYLSRLLTKRRIPHNVLNAKYHEQEAAIIAEAGRRGAVTVATNMAG
MAV_2894|M.avium_104 VAESEELHERLLRRGVPAVVLNAKNDAEEAQVIAEAGKFGVVTVSTQMAG
* .** * . : :* :* ***** . :** ::* **: * :**:*:***
Mflv_4705|M.gilvum_PYR-GCK RGTDIVLGGNVDYLLDRRLRQRGLDPIETPDEYEKGWHEELPHIKEQVAE
Mvan_1762|M.vanbaalenii_PYR-1 RGTDIVLGGNVDYLLDRRLRQRGLDPIETPEEYEQGWHEELPHIKAEVAA
MSMEG_1881|M.smegmatis_MC2_155 RGTDIVLGGNVDFLADKRLRERGLDPVETPEEYEAAWHEVLPQVKAECAK
Mb3268c|M.bovis_AF2122/97 RGTDIVLGGNVDFLTDQRLRERGLDPVETPEEYEAAWHSELPIVKEEASK
Rv3240c|M.tuberculosis_H37Rv RGTDIVLGGNVDFLTDQRLRERGLDPVETPEEYEAAWHSELPIVKEEASK
MLBr_00779|M.leprae_Br4923 RGTDIVLGGNVDFLTDQRLRRRGLDPVETPDEYEQAWHSELPKVKEEAGQ
MMAR_1305|M.marinum_M RGTDIVLGGNVDFLTDKRLRDNGLDPVETPDEYEQAWHQELPKVKEEAGD
MUL_2576|M.ulcerans_Agy99 RGTDIVLGGNVDFLTDKRLRDNGLDPVETPDEYEQAWHQELPKVKEEAGD
MAB_3580c|M.abscessus_ATCC_199 RGTDIVLGGNPDFLADKHLREQGLDPVETPDEYQAAWDETLQKFKDAAET
TH_0994|M.thermoresistible__bu RGTDIVLGGNPDFLTDKRLREQGLDPVETPEEYEAAWHELLPKVKAECAR
MAV_2894|M.avium_104 RGTDIRLGGSDEADHDR---------------------------------
***** ***. : *:
Mflv_4705|M.gilvum_PYR-GCK EAKDVIAAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLADE
Mvan_1762|M.vanbaalenii_PYR-1 EAKDVIAAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLADE
MSMEG_1881|M.smegmatis_MC2_155 EAEQVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
Mb3268c|M.bovis_AF2122/97 EAKEVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
Rv3240c|M.tuberculosis_H37Rv EAKEVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
MLBr_00779|M.leprae_Br4923 EAAEVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
MMAR_1305|M.marinum_M EATEVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
MUL_2576|M.ulcerans_Agy99 EATEVIKAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
MAB_3580c|M.abscessus_ATCC_199 EAKEVQEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLQDE
TH_0994|M.thermoresistible__bu EAQEVIEAGGLYVLGTERHESRRIDNQLRGRSGRQGDPGESRFYLSLGDE
MAV_2894|M.avium_104 ----VAELGGLHVVGTGRHHTERLDNQLRGRAGRQGDPGSSVFFSSWEDD
* ***:*:** **.:.*:*******:*******.* *: * *:
Mflv_4705|M.gilvum_PYR-GCK LMRRFNGATLETLLTRLNLPDDVPIEAKMVSRAIKSAQTQVEQQNFDIRK
Mvan_1762|M.vanbaalenii_PYR-1 LMRRFNGATLETLLTRLNLPDDVPIEAKMVSRAIKSAQTQVEQQNFDIRK
MSMEG_1881|M.smegmatis_MC2_155 LMRRFNGATLETLLTRLNLPDDVPIEAKMVSRAIKSAQTQVEQQNFEVRK
Mb3268c|M.bovis_AF2122/97 LMRRFNGAALETLLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
Rv3240c|M.tuberculosis_H37Rv LMRRFNGAALETLLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
MLBr_00779|M.leprae_Br4923 LMRRFHGATLEALLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
MMAR_1305|M.marinum_M LMRRFNGAALESLLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
MUL_2576|M.ulcerans_Agy99 LMRRFNGAALESLLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
MAB_3580c|M.abscessus_ATCC_199 LMRRFNGAALETILTRMNVPDDVPIEAKMVTNAIKSAQTQVEQQNFEVRK
TH_0994|M.thermoresistible__bu LMRRFNGATLEALLTRLNLPDDVPIEAKMVTRAIKSAQTQVEQQNFEVRK
MAV_2894|M.avium_104 VVAANLDRNKLPMETDPETGDGRIVSPKAAG-LLDHAQRVAEGRMLDVHA
:: . .: * : *. :..* . :. ** .* : ::::
Mflv_4705|M.gilvum_PYR-GCK EVLKYDEVMNQQRKVIYAERRRILEGENLRDQAEQMLVDVVTAYVDGATS
Mvan_1762|M.vanbaalenii_PYR-1 EVLKYDEVMNQQRKVVYAERRRILEGENLAGQAHQILVDVITAYVDGATA
MSMEG_1881|M.smegmatis_MC2_155 NVLKYDEVMNQQRKVIYAERRRILEGENLAEQAHKMLVDVITAYVDGATA
Mb3268c|M.bovis_AF2122/97 NVLKYDEVMNQQRKVIYAERRRILEGENLKDQALDMVRDVITAYVDGATG
Rv3240c|M.tuberculosis_H37Rv NVLKYDEVMNQQRKVIYAERRRILEGENLKDQALDMVRDVITAYVDGATG
MLBr_00779|M.leprae_Br4923 NVLKYDEVMNQQRKVIYAERRRILEGESLQQQALGMVRDVITAYIDGATT
MMAR_1305|M.marinum_M NVLKYDEVMNQQRKVIYAERRRILEGENLQQQAKDMLTDVITAYVDGATV
MUL_2576|M.ulcerans_Agy99 NVLKYDEVMNQQRKVIYAERRRILEGENLQQQVKDMLTDVITAYVDGATV
MAB_3580c|M.abscessus_ATCC_199 NVLKYDEVMNQQRKVIYAERRRILDGEDLQPQIQEMITDTIAAYVDGATA
TH_0994|M.thermoresistible__bu NVLKYDEVMNQQRKVIYAERRRILEGENLREQALDMVRDVITAYVDGATA
MAV_2894|M.avium_104 NTWRYNQLIAQQRAIIVDRRNTLLR---------------------TATA
:. :*:::: *** :: .*. :* **
Mflv_4705|M.gilvum_PYR-GCK EGYSEDWDLEKLWEGLRQLYPVGIDHRDLIDSDAIGEPGELTRAELLEAL
Mvan_1762|M.vanbaalenii_PYR-1 EGYSEDWDLEKLWEGLRQLYPVGIDHHDLIDSDAVGEPGELTREELLQAL
MSMEG_1881|M.smegmatis_MC2_155 EGYAEDWDLETLWTALKTLYPVGIDHRDLIDSDAVGEPGELTREELLDAL
Mb3268c|M.bovis_AF2122/97 EGYAEDWDLDALWTALKTLYPVGITADSLTRKDHEFERDDLTREELLEAL
Rv3240c|M.tuberculosis_H37Rv EGYAEDWDLDALWTALKTLYPVGITADSLTRKDHEFERDDLTREELLEAL
MLBr_00779|M.leprae_Br4923 DSYVEDWDLDALWSALGTLYPVGIKHESLTHADEDSECDELSRDELLEAL
MMAR_1305|M.marinum_M EGYAEDWDLDALWTALKTLYPVGIKADTLMRRDQDSDRDDLTRDELLEAL
MUL_2576|M.ulcerans_Agy99 EGYAEDWDLDALWTALKTLYPVGIKTDTLMRRDQDSDRDDLTRDELLQAL
MAB_3580c|M.abscessus_ATCC_199 DGYHEDWDFDALWTALKTLYPVSLKPEELIASGEYGEADELSPEDLKAAL
TH_0994|M.thermoresistible__bu EGYAEDWDLEQLWTALRQLYPISIDHHDLIDSDAVGEPGDLTRDELLQVL
MAV_2894|M.avium_104 R----------------------------------------------EEL
*
Mflv_4705|M.gilvum_PYR-GCK VNDAKSAYSVREAEIEQIAGEGAMRQLERNVLLNVLDRKWREHLYEMDYL
Mvan_1762|M.vanbaalenii_PYR-1 IADAERAYAAREAEIEEIAGEGAMRQLERNVLLNVLDRKWREHLYEMDYL
MSMEG_1881|M.smegmatis_MC2_155 IKDAERAYAEREKQIEAIAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
Mb3268c|M.bovis_AF2122/97 LKDAERAYAAREAELEEIAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
Rv3240c|M.tuberculosis_H37Rv LKDAERAYAAREAELEEIAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
MLBr_00779|M.leprae_Br4923 LIDAQHAYAAREAELAELAGEGAMRQLERNVLLNVVDRKWREHLYEMDYL
MMAR_1305|M.marinum_M LQDADQAYAAREAELEELAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
MUL_2576|M.ulcerans_Agy99 LQDADQAYAAREAELEELAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
MAB_3580c|M.abscessus_ATCC_199 LEDAKKAYKAREAEIDGLAGEGSMRQLERNILLSVIDRKWREHLYEMDYL
TH_0994|M.thermoresistible__bu LEDAERAYAKREAEIDELAGEGAMRQLERNVLLNVIDRKWREHLYEMDYL
MAV_2894|M.avium_104 AELAPKRYR----ELAEEIPEERLETICRHIMLYHLDRGWADHLAYLADI
* * :: * :. : *:::* :** * :** : :
Mflv_4705|M.gilvum_PYR-GCK REGIGLRGLAQQRPEVEYARE--GYDMFIGMLDGMKEESVGFLFNVQVEP
Mvan_1762|M.vanbaalenii_PYR-1 REGIGLRGLAQQRPEVEYARE--GYDMFIAMLDGMKEESVGFLFNVQVER
MSMEG_1881|M.smegmatis_MC2_155 KEGIGLRAMAQRDPLVEYQRE--GYDMFVGMLEALKEESVGFLFNVQVEA
Mb3268c|M.bovis_AF2122/97 KEGIGLRAMAQRDPLVEYQRE--GYDMFMAMLDGMKEESVGFLFNVTVEA
Rv3240c|M.tuberculosis_H37Rv KEGIGLRAMAQRDPLVEYQRE--GYDMFMAMLDGMKEESVGFLFNVTVEA
MLBr_00779|M.leprae_Br4923 KEGIGLRAMAQRDPLVEYQRE--GYDMFMAMLDGVKEESVGFLFNLAVEA
MMAR_1305|M.marinum_M KEGIGLRAMAQRDPLVEYQRE--GYDMFMAMLDGMKEESVGFLFNVSVEA
MUL_2576|M.ulcerans_Agy99 KEGIGLRAMAQRDPLVEYQRE--GYDMFMAMLDGVKEESVGFLFNVSVEA
MAB_3580c|M.abscessus_ATCC_199 KEGIGLRAMAQRDPLVEYQRE--GYDMFTAMLDGLKEESVGFLFNINVEV
TH_0994|M.thermoresistible__bu KEGIGLRAMAQRDPLVXXWRPRVGQSRALSRRQACLDPTSRGDEDLAVPV
MAV_2894|M.avium_104 RESIHLRALGRQNPLDEFHRL-----------------------------
:*.* **.:.:: * *
Mflv_4705|M.gilvum_PYR-GCK V-AAP-TVAAQAAPSGLAEFAAAAAQQ----------GSVATKERP----
Mvan_1762|M.vanbaalenii_PYR-1 APSAP-TVAAQAAPAGLAAFAAAAAEQA-----QAQTGGVATKERPA---
MSMEG_1881|M.smegmatis_MC2_155 APQQP-QVAPQAPPPTLSEFAAAAAAKASDSAAKPDSGSVATKERAEAER
Mb3268c|M.bovis_AF2122/97 VPAP--PVAPAAEPAELAEFAAAAAAAAQ-------QRSA--VDGGARER
Rv3240c|M.tuberculosis_H37Rv VPAP--PVAPAAEPAELAEFAAAAAAAAQ-------QRSA--VDGGARER
MLBr_00779|M.leprae_Br4923 VPAS--HVAPVEIPEGFTGLVADVEAIR------------------PREE
MMAR_1305|M.marinum_M VPAPQVEVAPVAEPEDLAEFATAAAAAAQ-------EGGAGPKTAAAREE
MUL_2576|M.ulcerans_Agy99 VPAPQVEVAPVAEPEDLAEFATAAAAAAQ-------EGGAGRKNAAAREE
MAB_3580c|M.abscessus_ATCC_199 QQPDGAAVAPQEAPEGLAEFASAAAQAAH-------G-------------
TH_0994|M.thermoresistible__bu GVCQFVERRGDAVQADLAGDQRRHIDVAVG---------------DRAQR
MAV_2894|M.avium_104 ------------AVDAFASLAADAIEAAQ---------------------
::
Mflv_4705|M.gilvum_PYR-GCK -TGGLRAKGIDDQSRPLTYTGPSEDGTPEVKRSENAP-----AASTGMTR
Mvan_1762|M.vanbaalenii_PYR-1 -VGGLRAKGIDDKAQPLTYTGPSEDGGVEVKRSGGG------TPSTGGTR
MSMEG_1881|M.smegmatis_MC2_155 PAPALRAKGIDNEAPPLTYTGPSEDGTAQVQRSGNGGR---HAAPAGGSR
Mb3268c|M.bovis_AF2122/97 APSALRAKGVASESPALTYSGPAEDGSAQVQRNGGGAHKTPAGVPAGASR
Rv3240c|M.tuberculosis_H37Rv APSALRAKGVASESPALTYSGPAEDGSAQVQRNGGGAHKTPAGVPAGASR
MLBr_00779|M.leprae_Br4923 ASSALRTKDIDNESTGLTYSGPSEDGSTQVQLNSGGGQKTPAGIPVGASR
MMAR_1305|M.marinum_M APSRLRAKGIEDESPALTYSGPSEDGSAQVQRNGGGAAKTPAGVPAGGSR
MUL_2576|M.ulcerans_Agy99 APSRLRAKGIEDESPALTYSGPSEDGSAQVQRNGGGAAKTPAGVPAGRSR
MAB_3580c|M.abscessus_ATCC_199 ---ELRAKGLEESTPELTYSGPAEDGSAQSRHENG-----AASASGGGTR
TH_0994|M.thermoresistible__bu FGELGRVVGVDEGDADLLGGAEERQHGVGFHAHPGDD----HPGPLWGPA
MAV_2894|M.avium_104 --QTFETANVLEDEPGLDLSKLAR--------------------PTSTWT
.. .: . * . .
Mflv_4705|M.gilvum_PYR-GCK KERREAARQQQKAGRHAKRG---
Mvan_1762|M.vanbaalenii_PYR-1 KERREAARQQ-KTGRHAKRR---
MSMEG_1881|M.smegmatis_MC2_155 RERREAARKQAKADRPAKSHRKG
Mb3268c|M.bovis_AF2122/97 RERREAARRQGRGAKPPKSVKKR
Rv3240c|M.tuberculosis_H37Rv RERREAARRQGRGAKPPKSVKKR
MLBr_00779|M.leprae_Br4923 RERREAARRRGRGAKPSRSVKKR
MMAR_1305|M.marinum_M RERREAARRQGRGAKPPKSVKKR
MUL_2576|M.ulcerans_Agy99 RERREAARRQGRGAKPPKSVKKR
MAB_3580c|M.abscessus_ATCC_199 RERREAARNAGRTSKPAKSRRKR
TH_0994|M.thermoresistible__bu QHIVEDAGHADRLEHHRRAGGER
MAV_2894|M.avium_104 YMVNDNPLSDDTLSTLSLPGVFR
: .