For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLTWEDDVEVHALRKRGWTISAIARHTGRDRKTIADYLSGKRTPGVRARPGPDPFEPFVDYVAARLTEDP HLWARTLLDELEQLGFPLSYQSLTRNIRARNLRPDCQACRTATERPNSVIEHAPGDETQWDWLELPDPPE SWGWGKTAHLLVGSLAHSGKWRAALTPSEDQPQLVAALDRVTRELGGCTRVWRFDRMATVCDPGSGRVTA SFAGVAKHYGVSVAICPPRRGNRKGVVEKVNHTAAQRWWRTLDDELTAEAAQASVDRFARVRGGTRMRAT ADGRSAVAVVAKTEPLAPVPATPYPVIISQTRTASRQALVSYRGNRYSVPPELAAAQVVVSHPVGAQFCD IATVAGIVIARHRMAADGLGVMVRDSGHVIALDTMAMATASTGRPHRRKERIPPGPAAKAAAAQLLHLND ANQEAAESSTWSTDSTVIDLSAYERAAQKRTIQ
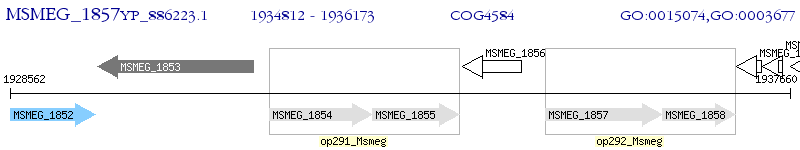
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1857 | - | - | 100% (453) | transposase IstA protein, putative |
| M. smegmatis MC2 155 | MSMEG_2151 | - | 0.0 | 100.00% (453) | transposase IstA protein, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0690 | - | 1e-121 | 81.60% (250) | transposase IstA protein, putative |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3774 | - | 1e-152 | 87.29% (291) | PUTATIVE transposase IstA protein, putative |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5931 | - | 1e-170 | 70.38% (395) | transposase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1857|M.smegmatis_MC2_155 MLTWEDDVEVHALRKRGWTISAIARHTGRDRKTIADYLSGKRTPGVRARP
Mvan_5931|M.vanbaalenii_PYR-1 MLTWEDDLEIHALHKRGWSISAIARHTGKNRRTVRNYLNGVTTPGVRKPA
Mflv_0690|M.gilvum_PYR-GCK MLTWEDDVEVHALRKRGWSISAIARHTKFDRKTVRKYLAGDGTPGVRIRT
TH_3774|M.thermoresistible__bu MLTWEDDVEVHALAKRGWTISAIARHTGFDRKTVRKYLAGDGTPGVRARP
*******:*:*** ****:******** :*:*: .** * ***** .
MSMEG_1857|M.smegmatis_MC2_155 GPDPFEPFVDYVAARLTEDPHLWARTLLDELEQLGFPLSYQSLTRNIRAR
Mvan_5931|M.vanbaalenii_PYR-1 AIDPFEPFIAYVTARLTEDPHLWARTLCDELEDLGYPMSYPTLTRQIRAR
Mflv_0690|M.gilvum_PYR-GCK GPDPFEPFVDYVVARLSEDPHLWARTLYDELEDLGFGLSYQSLTRHLRDR
TH_3774|M.thermoresistible__bu GPDPFDPFVDYVTARLTEDPHLWARTLFDELEDLGFGLSYQSLTRNIRTR
. ***:**: **.***:********** ****:**: :** :***::* *
MSMEG_1857|M.smegmatis_MC2_155 NLRPDCQACRTATERPNSVIEHAPGDETQWDWLELPDPPESWGWGKTAHL
Mvan_5931|M.vanbaalenii_PYR-1 NLRPACQDCAHAADRANAVIDHPPGSETQWDWLDLPNPPASWGWGTMAHL
Mflv_0690|M.gilvum_PYR-GCK KLRPVCAACRTATQRPNAVIAHAPGDETQWDWLELPDPPDSWGWGKTAHL
TH_3774|M.thermoresistible__bu NLRPVCEACRTATGRPNAVIPHPPGEETQWDWLELPDPPASWGWGKTAHL
:*** * * *: *.*:** *.**.*******:**:** *****. ***
MSMEG_1857|M.smegmatis_MC2_155 LVGSLAHSGKWRAALTPSEDQPQLVAALDRVTRELGGCTRVWRFDRMATV
Mvan_5931|M.vanbaalenii_PYR-1 FVGSLAHSGRWRAVLAPAMTQPHLVDGLDRISRKLGGLTRTWRFDRMATV
Mflv_0690|M.gilvum_PYR-GCK LVGSLAHSGKWRGALASSEDQPHLVACLDRVTRGLGGVTRVWRFDRMATV
TH_3774|M.thermoresistible__bu LVGSLSHSGKWRGNLAPSEDQPHLVAGLDRVTRGLGGLTRVWRFDRMATV
:****:***:**. *:.: **:** ***::* *** **.*********
MSMEG_1857|M.smegmatis_MC2_155 CDPGSGRVTASFAGVAKHYGVSVAICPPRRGNRKGVVEKVNHTAAQRWWR
Mvan_5931|M.vanbaalenii_PYR-1 CDPGSGRVTASFAGVAKHYGVAVAICPARRGNRKGVVEKANHTAAQRWWR
Mflv_0690|M.gilvum_PYR-GCK CDPGSGRVTASFAGVAKHYGVSVAICPPRRGNRK------------AWWR
TH_3774|M.thermoresistible__bu CDPGSGRVTASFAGVAKHYGVSVAICPPRRGNRKGVVEKVNHTAAQRWWR
*********************:*****.****** ***
MSMEG_1857|M.smegmatis_MC2_155 TLDDELTAEAAQASVDRFARVRGGTRMRATADGRSAVAVVAKT-------
Mvan_5931|M.vanbaalenii_PYR-1 TLPDDITVEQAQDRLDEFCRQRGDARRRATAEGKVTVATLAAA-------
Mflv_0690|M.gilvum_PYR-GCK ---------------------------------RSTTPPPN---------
TH_3774|M.thermoresistible__bu TLADDLTVEAAQASLDRFARVRGDTRLRATADGRSSVAVVAXIYVRTAEQ
: :..
MSMEG_1857|M.smegmatis_MC2_155 -----------EPLAP------------VPATP--------YPVIISQTR
Mvan_5931|M.vanbaalenii_PYR-1 -----------EPLAP------------LPVAP--------YPLILREDR
Mflv_0690|M.gilvum_PYR-GCK -----------AGGAP-----------WPTT-------------------
TH_3774|M.thermoresistible__bu WVGVCPGCMVRSGVCPGCMVRSARSKGWVSTRPRDIKIGPDIPRIVWRKR
.* ..
MSMEG_1857|M.smegmatis_MC2_155 -------------------TASRQALVSYRGNR-----------------
Mvan_5931|M.vanbaalenii_PYR-1 -------------------TVSRQAMVSYRGNR-----------------
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu KWVCANMCCDRKSFTESVPSIPPRARVTVRAKREMASAVLDDDRSVKAVA
MSMEG_1857|M.smegmatis_MC2_155 ---------------YSVPPELAAAQVVVSHP------------------
Mvan_5931|M.vanbaalenii_PYR-1 ---------------YSVPPELASATVSVIHV------------------
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu AAYGCTWNTCHDAVIATADPVLAGEPEAVSVLGIDETRRGKAKWETCSQT
MSMEG_1857|M.smegmatis_MC2_155 --------VGAQFCDIATVAGIVIARHRMAADGLG--VMVRDSGHVIALD
Mvan_5931|M.vanbaalenii_PYR-1 --------LGSEVIDIVTSSQITIARHRLAADGAG--VMVRDHGHVVALD
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu GTRSWVDRFDTGLVDITGTGGLLVQVNGRAAKPVTDWLDQRDPGWKATIE
MSMEG_1857|M.smegmatis_MC2_155 TMAMATASTGRPHRRKERIPPGPAAKAAAAQLLHLNDANQEAAESSTWST
Mvan_5931|M.vanbaalenii_PYR-1 QLAMATAATGGPSS-----PPQ--------------------------GT
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu FVAIDMSVTYANAAR--QALPHAKLIVDRFHLVKRANEMVDEVRRRTTQA
MSMEG_1857|M.smegmatis_MC2_155 DSTVIDLSAYERAAQKRTIQ------------------------------
Mvan_5931|M.vanbaalenii_PYR-1 HPTRRSIARSSRSIAHQRI-------------------------------
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu HRLRRGRKSDPEWINRRRLLRAAERLTDEQRHKLFVKLTSADPNGDIAAA
MSMEG_1857|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5931|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu WIAKELLRDVLACTARGGLTYEIRDALYRFYTFCAACSVPEISKLARTIS
MSMEG_1857|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5931|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0690|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu AWQEPMILAIQTGLSNARSEGYNRIIKHVGRIAFGFRNPENQRRRVRWAC
MSMEG_1857|M.smegmatis_MC2_155 -----------------
Mvan_5931|M.vanbaalenii_PYR-1 -----------------
Mflv_0690|M.gilvum_PYR-GCK -----------------
TH_3774|M.thermoresistible__bu TRQSRRTPPSTKQVRPC