For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMEGMSTPLSLEKITNAPKALLHDHLDGGLRPATVLDLAGQVGYDNLPATDADELATFFRTAAHSGSLVR YLEPFAHTVGVMQTPEALHRVAFECVEDLAADNVVYAEVRFAPELHIDGGLSLDDVVDAVLAGFADGEKA SAAAGRTIVVRCLVTAMRHAARSREIAELAIRFRDKGVVGFDIAGAEAGYPPSRHLDAFEYMRSNNARFT IHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDISETPDGTQQLGRLASLLRDKRIPLEMCPSSNVQTGA VKSIAEHPFDRLARLRFRVTVNTDNRLMSDTTMSQEMHRLVEAFGYGWSDLERFTINAMKSAFIPFDQRL AIIDEVIKPRYAVLVG
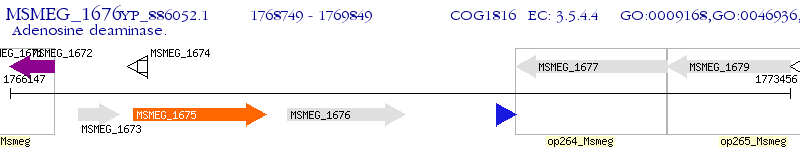
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1676 | add | - | 100% (366) | adenosine deaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3342c | add | 1e-174 | 83.70% (362) | adenosine deaminase |
| M. gilvum PYR-GCK | Mflv_4841 | - | 0.0 | 88.67% (362) | adenosine deaminase |
| M. tuberculosis H37Rv | Rv3313c | add | 1e-174 | 83.70% (362) | adenosine deaminase |
| M. leprae Br4923 | MLBr_00700 | add | 1e-178 | 85.36% (362) | adenosine deaminase |
| M. abscessus ATCC 19977 | MAB_3670c | - | 1e-170 | 81.49% (362) | adenosine deaminase |
| M. marinum M | MMAR_1206 | add | 1e-172 | 83.47% (357) | adenosine deaminase Add |
| M. avium 104 | MAV_4294 | add | 1e-176 | 83.33% (366) | adenosine deaminase |
| M. thermoresistible (build 8) | TH_1646 | add | 0.0 | 84.53% (362) | PROBABLE ADENOSINE DEAMINASE ADD (ADENOSINE AMINOHYDROLASE) |
| M. ulcerans Agy99 | MUL_1364 | add | 1e-171 | 83.19% (357) | adenosine deaminase |
| M. vanbaalenii PYR-1 | Mvan_1592 | - | 0.0 | 89.50% (362) | adenosine deaminase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4841|M.gilvum_PYR-GCK ----MTTPLTLENIRRAPKALLHDHLDGGLRPSTVLELAEQYGYDDLPAH
Mvan_1592|M.vanbaalenii_PYR-1 ----MTTPLTFDNIRRAPKALLHDHLDGGLRPSTVLELAEQYGYEDLPAH
MSMEG_1676|M.smegmatis_MC2_155 MMEGMSTPLSLEKITNAPKALLHDHLDGGLRPATVLDLAGQVGYDNLPAT
MMAR_1206|M.marinum_M ----MTDMPTLDAIRQAPKALLHDHLDGGLRPETVLDIAGQVGYDGLPST
MUL_1364|M.ulcerans_Agy99 ----MTDMPTLDAIRQAPKALLHDHLDGGLRPETVLDIAGQVGYDGLPST
MAV_4294|M.avium_104 ----MTAPLELEQIRKAPKALLHDHLDGGLRPSTVLDIAGQTGYDGLPAT
Mb3342c|M.bovis_AF2122/97 ----MTAAPTLQTIRLAPKALLHDHLDGGLRPATVLDIAGQVGYDDLPAT
Rv3313c|M.tuberculosis_H37Rv ----MTAAPTLQTIRLAPKALLHDHLDGGLRPATVLDIAGQVGYDDLPAT
MLBr_00700|M.leprae_Br4923 ----MNTPLHLENIKQAPKALLHDHLDGGLRPATVLDIAGQVGYDRLPAT
TH_1646|M.thermoresistible__bu ----MTTPLTLENIAKAPKALLHDHLDGGLRPATVIELAEETGYDDLPTT
MAB_3670c|M.abscessus_ATCC_199 ----MTAELDLVSITQAPKALLHDHLDGGLRPGTVLDLARETGYENLPAQ
*. : * **************** **:::* : **: **:
Mflv_4841|M.gilvum_PYR-GCK DADELAEFFRTAAHSGSLVRYLEPFAHTVGVMQNHDALHRVARECVEDLA
Mvan_1592|M.vanbaalenii_PYR-1 DADGLATFFRTAAHSGSLVRYLEPFAHTVGVMQNPDALHRVARECVEDLA
MSMEG_1676|M.smegmatis_MC2_155 DADELATFFRTAAHSGSLVRYLEPFAHTVGVMQTPEALHRVAFECVEDLA
MMAR_1206|M.marinum_M DAGELASWFRTQSHSGSLERYLEPFSHTVAVMQTPEALYRVAYECVEDLA
MUL_1364|M.ulcerans_Agy99 DAGELASWFRTQSHSGSLERYLEPFSHTVAVMQTPEALYRVAYECVEDLA
MAV_4294|M.avium_104 DVEELATWFRTRSHSGSLERYLEPFSHTVAVMQTPEALHRVAYECVEDLA
Mb3342c|M.bovis_AF2122/97 DVDALASWFRTQSHSGSLERYLEPFSHTVAVMQTPEALYRVAFECAQDLA
Rv3313c|M.tuberculosis_H37Rv DVDALASWFRTQSHSGSLERYLEPFSHTVAVMQTPEALYRVAFECAQDLA
MLBr_00700|M.leprae_Br4923 DVESLETWFRTASHSGSLERYLEPFSHTVAVMQTPEALHRVAYECVEDLA
TH_1646|M.thermoresistible__bu DVDELADWFRTAAHSGSLERYLEPFAHTVGVLQTPEALHRVAYECVEDLA
MAB_3670c|M.abscessus_ATCC_199 DETALASWFRTAADSGSLEQYLETFAHTVGVMQTASALHRVAAECVEDLA
* * :*** :.**** :***.*:***.*:*. .**:*** **.:***
Mflv_4841|M.gilvum_PYR-GCK DDNVVYAEIRFAPELHIDGGLSLDAVVEAVLAGFADGEKAAAAAGRTITV
Mvan_1592|M.vanbaalenii_PYR-1 ADNVVYAEVRFAPELHIDGGLSLDAVVDAVLAGFADGEKAAAADGRAITV
MSMEG_1676|M.smegmatis_MC2_155 ADNVVYAEVRFAPELHIDGGLSLDDVVDAVLAGFADGEKASAAAGRTIVV
MMAR_1206|M.marinum_M ADAVVYAEIRFAPELHINQGLTFDEIVDAVLAGFAAGERACAGAGCPIKV
MUL_1364|M.ulcerans_Agy99 ADAVVYAEIRFAPELHINRGLTFDEIVDAVLAGFAAGERACAGAGCPIKV
MAV_4294|M.avium_104 EDSVVYAEIRFAPELHINRGMSFDEIVDAVLAGFADGEKACAAAGRPIVV
Mb3342c|M.bovis_AF2122/97 ADSVVYAEVRFAPELHISCGLSFDDVVDTVLTGFAAGEKACAADGQPITV
Rv3313c|M.tuberculosis_H37Rv ADSVVYAEVRFAPELHISCGLSFDDVVDTVLTGFAAGEKACAADGQPITV
MLBr_00700|M.leprae_Br4923 ADSVVYAEVRFAPELHIDEGLSFDEVLASVLAGFADGERACAAEGNAITV
TH_1646|M.thermoresistible__bu ADNVVYAEVRFAPELHIDGGMSLDEVVEAVLAGFAEGEKAAAAEGRTIMV
MAB_3670c|M.abscessus_ATCC_199 ADGVVYAEVRFAPELHIDAGLTLDDVMDAVLAGFADGERQASAAGKRIKV
* *****:********. *:::* :: :**:*** **: .:. * * *
Mflv_4841|M.gilvum_PYR-GCK RCLVTAMRHAARSREIAALAIRFRDQGVVGFDIAGAEAGYPPSRHLDAFE
Mvan_1592|M.vanbaalenii_PYR-1 RCLVTAMRHAARSREIAELAIRFRDKGVVGFDIAGAEAGYPPSRHLDAFE
MSMEG_1676|M.smegmatis_MC2_155 RCLVTAMRHAARSREIAELAIRFRDKGVVGFDIAGAEAGYPPSRHLDAFE
MMAR_1206|M.marinum_M RLLVTAMRHAAMSREIAELAIRFRDKGVVGFDIAGAEAGYPPSRHLDAFE
MUL_1364|M.ulcerans_Agy99 RLLVTAMRHAAMSREIAELAIRFRDKGVVGFDIAGAEAGYPPSRHLDAFE
MAV_4294|M.avium_104 RLLVTAMRHAAVSREIAELAIRWRDKGVVGFDIAGAEAGNPPTRHLEAFD
Mb3342c|M.bovis_AF2122/97 RCLVTAMRHAAMSREIAELAIRFRDKGVVGFDIAGAEAGHPPTRHLDAFE
Rv3313c|M.tuberculosis_H37Rv RCLVTAMRHAAMSREIAELAIRFRDKGVVGFDIAGAEAGHPPTRHLDAFE
MLBr_00700|M.leprae_Br4923 RCLVTAMRHAAMSREIAELAIRFRDKGVVGFDIAGAEAGHPPTRHLDAFE
TH_1646|M.thermoresistible__bu RCLVTAMRHAARSREIAELAIRYRDDGVVGFDIAGAEAGHPPTRHLDAFE
MAB_3670c|M.abscessus_ATCC_199 RTLVTAMRHAARSREIAELAVKFRDRGAVGFDIAGAEAGYPPTRHLDAFE
* ********* ***** **:::** *.*********** **:***:**:
Mflv_4841|M.gilvum_PYR-GCK YMRSNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIDMDAEG
Mvan_1592|M.vanbaalenii_PYR-1 YMRSNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIEIDADG
MSMEG_1676|M.smegmatis_MC2_155 YMRSNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDISETPDG
MMAR_1206|M.marinum_M YMRDNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIEVGPDG
MUL_1364|M.ulcerans_Agy99 YMRDNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIEVGLDG
MAV_4294|M.avium_104 YMRDHNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIDVLADG
Mb3342c|M.bovis_AF2122/97 YMRDHNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIDVDADG
Rv3313c|M.tuberculosis_H37Rv YMRDHNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIDVDADG
MLBr_00700|M.leprae_Br4923 YMRSNNARFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIDVDPGG
TH_1646|M.thermoresistible__bu FMRGNNGRFTIHAGEAFGLPSIHEAIAFCGADRLGHGVRIVDDIEQRPDG
MAB_3670c|M.abscessus_ATCC_199 YMRNANSRFTIHAGEGFGLPSIHEAIAFCGADRLGHGVRIVDDIDIGPDG
:**. *.********.****************************. *
Mflv_4841|M.gilvum_PYR-GCK ----GPKLGRLAALLRDKRIPFEMCPSSNVQTGAVASIAEHPFDRLARLR
Mvan_1592|M.vanbaalenii_PYR-1 ----NAKLGRLASLLRDKRIPFEMCPSSNVQTGAVASIAEHPFDRLARLR
MSMEG_1676|M.smegmatis_MC2_155 ----TQQLGRLASLLRDKRIPLEMCPSSNVQTGAVKSIAEHPFDRLARLR
MMAR_1206|M.marinum_M ----DVKLGRLAAILRDKRIPLELCPSSNVQTGAVASIAEHPFDLLARSR
MUL_1364|M.ulcerans_Agy99 ----DVKLGRLAAILRDKRIPLELCPSSNVQTGAVASIAEHPFDLLARSR
MAV_4294|M.avium_104 PDKGKVRLGRLANILRDKRIPLELCPSSNVQTGAVKSIADHPFDLLARTR
Mb3342c|M.bovis_AF2122/97 ----GFQLGRLAAILRDKRIPLELCPSSNVQTGAVASIAEHPFDLLARAR
Rv3313c|M.tuberculosis_H37Rv ----GFQLGRLAAILRDKRIPLELCPSSNVQTGAVASIAEHPFDLLARAR
MLBr_00700|M.leprae_Br4923 ----GIRLGPLASILRDKRIPLELCPSSNLQTGAVASITEHPFDLLAWAR
TH_1646|M.thermoresistible__bu ----SYHLGRVASIVRDKRIPLEMCPTSNVQTGAVRSLAEHPFDILARLR
MAB_3670c|M.abscessus_ATCC_199 ----TAHLGRLSSLLRDKRVPLELCPSSNVQTGAVESLADHPFDLLARLR
:** :: ::****:*:*:**:**:***** *:::**** ** *
Mflv_4841|M.gilvum_PYR-GCK FRVTVNTDNRLMSDTTMSLEMLRLVEAFGYGWSDLERFTINAMKSAFISF
Mvan_1592|M.vanbaalenii_PYR-1 FRVTVNTDNRLMSDTSMSMEMLRLVEAFGYGWSDLERFTINAMKSAFIAF
MSMEG_1676|M.smegmatis_MC2_155 FRVTVNTDNRLMSDTTMSQEMHRLVEAFGYGWSDLERFTINAMKSAFIPF
MMAR_1206|M.marinum_M FRVTVNTDNRLMSDTSMSQEMYRLVETFGYGWSDIQRFTINAMKSAFIAF
MUL_1364|M.ulcerans_Agy99 FRVTVNTDNRLMSDTSMSQEMYRLVETFGYGWSDIQRFTINAMKSAFIAF
MAV_4294|M.avium_104 FRVTVNTDNRLMSDTYMSREMHRLVQAFGYGWSDLERFTINAMKSAFIPF
Mb3342c|M.bovis_AF2122/97 FRVTVNTDNRLMSDTSMSLEMHRLVEAFGYGWSDLARFTVNAMKSAFIPF
Rv3313c|M.tuberculosis_H37Rv FRVTVNTDNRLMSDTSMSLEMHRLVEAFGYGWSDLARFTVNAMKSAFIPF
MLBr_00700|M.leprae_Br4923 FRVTVNTDNRLMSDTSMSLEMHRLVEAFGYGWGDLERFTINAMKSAFIPF
TH_1646|M.thermoresistible__bu FRVTVNTDNRLMSDTSMSREMMRLVETFGYGWSDLQRFTINAMKSAFIPF
MAB_3670c|M.abscessus_ATCC_199 FRVTVNTDNRLMSDTTMSREMLRLVETFGYGWSDLQRFTINAMKSAFIPF
*************** ** ** ***::*****.*: ***:********.*
Mflv_4841|M.gilvum_PYR-GCK PERLAIIDEVIKPRYAVLVG---
Mvan_1592|M.vanbaalenii_PYR-1 DERLAIIDEVIKPRYAVLVG---
MSMEG_1676|M.smegmatis_MC2_155 DQRLAIIDEVIKPRYAVLVG---
MMAR_1206|M.marinum_M DERLEIIDEVIKPRFAVLIG---
MUL_1364|M.ulcerans_Agy99 DERLEIIDEVIKPRFAVLIG---
MAV_4294|M.avium_104 DERLAIIDEVIKPRYAVLIG---
Mb3342c|M.bovis_AF2122/97 DQRLAIIDEVIKPRFAALMGHSE
Rv3313c|M.tuberculosis_H37Rv DQRLAIIDEVIKPRFAALMGHSE
MLBr_00700|M.leprae_Br4923 DQRLAIIDEVIKPRFAVLVG---
TH_1646|M.thermoresistible__bu PDRLTLIDDVIKPRYAVLIG---
MAB_3670c|M.abscessus_ATCC_199 DERLAIIDDVIKPGYAVLIG---
:** :**:**** :*.*:*