For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDELERTDEPNLVRPYTLTAGRTAPPVNLPLEAPVETLDTPNAPTWPGRDVRGQIVELCTGSPSVAEIAA HLSLPLGVARVLIGDLVKQGYLRVHTTLEDSASFDERRELIGRTLRGLRAL
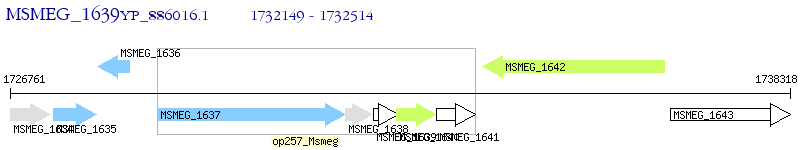
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1639 | - | - | 100% (121) | hypothetical protein MSMEG_1639 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3398c | - | 4e-38 | 68.18% (110) | hypothetical protein Mb3398c |
| M. gilvum PYR-GCK | Mflv_4887 | - | 8e-39 | 68.64% (118) | hypothetical protein Mflv_4887 |
| M. tuberculosis H37Rv | Rv3363c | - | 4e-38 | 68.18% (110) | hypothetical protein Rv3363c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1167 | - | 3e-35 | 62.39% (117) | hypothetical protein MMAR_1167 |
| M. avium 104 | MAV_4329 | - | 1e-37 | 67.57% (111) | hypothetical protein MAV_4329 |
| M. thermoresistible (build 8) | TH_0747 | - | 3e-39 | 72.81% (114) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2848 | - | 3e-27 | 64.84% (91) | hypothetical protein MUL_2848 |
| M. vanbaalenii PYR-1 | Mvan_1538 | - | 1e-44 | 76.99% (113) | hypothetical protein Mvan_1538 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1639|M.smegmatis_MC2_155 -------MDELER----TDEPNLVRPYTLTAGRTAPPVNLPLEAPVETLD
TH_0747|M.thermoresistible__bu -------MDAWQPEPGPADEPSLVRPYILTAGRTDSRVDLPLEAPIQAVP
Mflv_4887|M.gilvum_PYR-GCK -------MGAH---ERKDPAPSLVRPYTLTSGRTESGVTLPLEAPVGLAK
Mvan_1538|M.vanbaalenii_PYR-1 -------MGAHGKPERESAEPSLVRPYTLTAGRTASTVRLPLEAPVGVVS
Mb3398c|M.bovis_AF2122/97 -------MFNPA---GDRPKAGLVRPYTLTAGRTGTDVDLPLQAPVQTLP
Rv3363c|M.tuberculosis_H37Rv -------MFNPA---GDRPKAGLVRPYTLTAGRTGTDVDLPLQAPVQTLP
MMAR_1167|M.marinum_M MDRRDPPLAAPE---AAR-EASLVRPYALTAGRTGTDIDLPLEAPVQTSR
MUL_2848|M.ulcerans_Agy99 -----------------------------------------MEAPVQTSR
MAV_4329|M.avium_104 -------MDTPET-WPTSRSGNLVRPYTLTSGRTDTTVDLPVEAPIQTLQ
::**:
MSMEG_1639|M.smegmatis_MC2_155 TPNAPTWPGRDVRGQIVELCTGSPSVAEIAAHLSLPLGVARVLIGDLVKQ
TH_0747|M.thermoresistible__bu TQAPVHWPRSDVRGQIVELGARSLSVAEIAAHLSLPLGVARVLIGDLVTQ
Mflv_4887|M.gilvum_PYR-GCK SAPPPGWPAGDVRSRIVALCDARPSVAEIAAHLSVPLGVARVLVGDLVLQ
Mvan_1538|M.vanbaalenii_PYR-1 PQQPPRWPGSDVRGQILELCAERPSVAEIAAHLSLPLGVARVLVGDLVVQ
Mb3398c|M.bovis_AF2122/97 AGPAGRWPAYDMRRRILQLCIGSPSVAEISARLDLPVGVARVLVGDLVTS
Rv3363c|M.tuberculosis_H37Rv AGPAGRWPAYDMRRRILQLCIGSPSVAEISARLDLPVGVARVLVGDLVTS
MMAR_1167|M.marinum_M AGLARRWQPSDVRGRIVQLCLTCPSVAEISARLDLPVGVARVLVGDLVTS
MUL_2848|M.ulcerans_Agy99 AGLARRWQPSDVRGRIVQLCLTSPSVAEISARLDLPVGVARVLVGDLVTS
MAV_4329|M.avium_104 AGLTHRWPPGDARGRIVRLCLDSPSVAEISARLDLPLGVARVLVGDLVLS
. . * * * :*: * *****:*:*.:*:******:**** .
MSMEG_1639|M.smegmatis_MC2_155 GYLRVHTTLEDSASFDERRELIGRTLRGLRAL
TH_0747|M.thermoresistible__bu GYLRVQSTLGAVSTDDERRELIGRTLRGLRAL
Mflv_4887|M.gilvum_PYR-GCK GYLQVHATLGEAATPDERRALIGRTLRGLRAL
Mvan_1538|M.vanbaalenii_PYR-1 GYVRVHTTLGEVATADERRELIGRTLRGLRAL
Mb3398c|M.bovis_AF2122/97 GYLRVHATLTDRSTRDERHELIGRTLRGLKAL
Rv3363c|M.tuberculosis_H37Rv GYLRVHATLTDRSTRDERHELIGRTLRGLKAL
MMAR_1167|M.marinum_M DYLRVHATLTERSTRDERHELIGRTLRGLKAL
MUL_2848|M.ulcerans_Agy99 DYLRVHATLTERSTRDERHELIGRTLRGLKAL
MAV_4329|M.avium_104 GYLRVHKTLSERSTRDERHELIGRTLRGLRAL
.*::*: ** :: ***: *********:**