For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNLERLAHTLQITELLYRYAELVDAGDFDGVGQLLGRGAFMGVTGADAIAALFAATTRRFPEHGNRPRTR HLVLNPIIDIDGERARARSTFVVIQNTETVSLQPIVAGRYADMFALDDTGWYFTERRVDVEMIGDVSAHL MLDPDTLPK
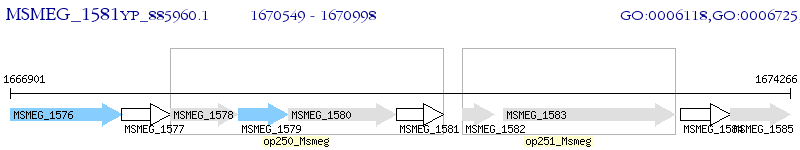
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1581 | - | - | 100% (149) | hypothetical protein MSMEG_1581 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4928 | - | 2e-58 | 76.64% (137) | hypothetical protein Mflv_4928 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3733c | - | 2e-48 | 63.70% (146) | hypothetical protein MAB_3733c |
| M. marinum M | MMAR_0323 | - | 5e-19 | 38.89% (144) | dioxygenase beta subunit |
| M. avium 104 | MAV_3825 | - | 8e-21 | 39.01% (141) | hypothetical protein MAV_3825 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4799 | - | 4e-19 | 38.89% (144) | dioxygenase beta subunit |
| M. vanbaalenii PYR-1 | Mvan_1490 | - | 5e-59 | 73.10% (145) | hypothetical protein Mvan_1490 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4928|M.gilvum_PYR-GCK MS---TADKLAVTELLYRYAELIDAGDFDGVGSLLGRGDFMG--------
Mvan_1490|M.vanbaalenii_PYR-1 MSGQ-LADKLAVTELLYRYAELIDAGDFDGVGELLGRGDFMG--------
MSMEG_1581|M.smegmatis_MC2_155 MNLERLAHTLQITELLYRYAELVDAGDFDGVGQLLGRGAFMG--------
MAB_3733c|M.abscessus_ATCC_199 MS---FADRLAITDLLYRYAELMDAGDFDGIGELLGRGSFGGEQG-----
MMAR_0323|M.marinum_M -----MRAETQIANLLYRYAECIDAGDLPAAAALFEHARIRIAADGDQDG
MUL_4799|M.ulcerans_Agy99 -----MRAETQIANLLYRYAECIDAGDLPAAAALFEHARIRIAADGDQDG
MAV_3825|M.avium_104 ----MTDSATEITNLIYTYAQLLDGGDLDGVARLFEHGRICGVQDGPPE-
:::*:* **: :*.**: . . *: :. :
Mflv_4928|M.gilvum_PYR-GCK ---VAGAEAISTLFAATTRRFPDHGNTPRTRHLVLNPIVDVDGE--VASA
Mvan_1490|M.vanbaalenii_PYR-1 ---VTGAEAIAGRFAATTRRFPEHGNTPRTRHLVLNPIVDVDGE--TATA
MSMEG_1581|M.smegmatis_MC2_155 ---VTGADAIAALFAATTRRFPEHGNRPRTRHLVLNPIIDIDGE--RARA
MAB_3733c|M.abscessus_ATCC_199 --SVSGASAITKLFFTTTRRYRETGGTPRTRHLVLNPVVEVAGDG-TASA
MMAR_0323|M.marinum_M GQDTVDAAGLLKIWEALIIVYPN--GTPRTKHVVSNPIIEIDPQCGIARC
MUL_4799|M.ulcerans_Agy99 GQDTVDAAGLLKIWEALIIVYPN--GTPRTKHVVSNPIIEIDPQCGIARC
MAV_3825|M.avium_104 -TVFAGSARVRQMYEVATRIYED--GTPKTKHNTTNVQLDIDEERGTARS
.: : : . : : . *:*:* . * ::: : * .
Mflv_4928|M.gilvum_PYR-GCK RSTFCVVQRTETVALQPIVVGRYRDTFARDTDGWHFTSRTV-DVEMIGDV
Mvan_1490|M.vanbaalenii_PYR-1 RSTFCVVQRTDNVPLQPIAVGRYADTFARDAGGWYFTSRAV-KVEMFGDV
MSMEG_1581|M.smegmatis_MC2_155 RSTFVVIQNTETVSLQPIVAGRYADMFALDDTGWYFTERRV-DVEMIGDV
MAB_3733c|M.abscessus_ATCC_199 RSTFCVVQATEQLPLQPIVVGRYRDTFGQDGHGWYFSSRRV-DVEMVGDV
MMAR_0323|M.marinum_M RSYYTVFQQVDDFPLQPIVTGRYHDRFACVDGKWRFSYRDLTLIDMVGDV
MUL_4799|M.ulcerans_Agy99 RSYYTVFQQVDDFPLQPIVTGRYHDRFACVDGKWRFSYRDLTLIDMVGDV
MAV_3825|M.avium_104 TSYYCVTQATPELPLQVIVTGHYQDTFHRVDGAWCFESRTM-FVDQVGDV
* : * * . ..** *..*:* * * * * * : :: .***
Mflv_4928|M.gilvum_PYR-GCK SDHLMMDLSRLG-----------
Mvan_1490|M.vanbaalenii_PYR-1 SEHLLIDPSRLSR----------
MSMEG_1581|M.smegmatis_MC2_155 SAHLMLDPDTLPK----------
MAB_3733c|M.abscessus_ATCC_199 SAHLLMDPGALSG----------
MMAR_0323|M.marinum_M SHHLRYALEPGPSTSANNRRSGP
MUL_4799|M.ulcerans_Agy99 SHHLRYALEPGPSTSANNRRSGP
MAV_3825|M.avium_104 SHHLKF-----------------
* **