For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKVHKGDTVLVISGKDKGAKGKVLVAYPDRNKVLVEGVNRIKKHTAVSANERGASSGGIVTQEAPIHVSN VMVVDSDGKPTRVGYRIDDETGKKVRIAKTNGKDI
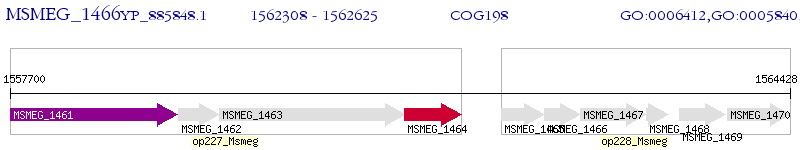
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1466 | rplX | - | 100% (105) | 50S ribosomal protein L24 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0736 | rplX | 1e-50 | 87.62% (105) | 50S ribosomal protein L24 |
| M. gilvum PYR-GCK | Mflv_5036 | rplX | 8e-49 | 88.57% (105) | 50S ribosomal protein L24 |
| M. tuberculosis H37Rv | Rv0715 | rplX | 1e-50 | 87.62% (105) | 50S ribosomal protein L24 |
| M. leprae Br4923 | MLBr_01848 | rplX | 8e-50 | 85.71% (105) | 50S ribosomal protein L24 |
| M. abscessus ATCC 19977 | MAB_3806c | rplX | 1e-49 | 84.76% (105) | 50S ribosomal protein L24 |
| M. marinum M | MMAR_1046 | rplX | 9e-51 | 86.67% (105) | 50S ribosomal protein L24, RplX |
| M. avium 104 | MAV_4454 | rplX | 7e-49 | 85.71% (105) | 50S ribosomal protein L24 |
| M. thermoresistible (build 8) | TH_0312 | rplX | 1e-51 | 92.38% (105) | PROBABLE 50S RIBOSOMAL PROTEIN L24 RPLX |
| M. ulcerans Agy99 | MUL_0805 | rplX | 7e-51 | 86.67% (105) | 50S ribosomal protein L24 |
| M. vanbaalenii PYR-1 | Mvan_1333 | rplX | 8e-49 | 89.52% (105) | 50S ribosomal protein L24 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0736|M.bovis_AF2122/97 MKVHKGDTVLVISGKDKGAKGKVLQAYPDRNRVLVEGVNRIKKHTAISTT
Rv0715|M.tuberculosis_H37Rv MKVHKGDTVLVISGKDKGAKGKVLQAYPDRNRVLVEGVNRIKKHTAISTT
MMAR_1046|M.marinum_M MKVHKGDTVLVVSGKDKGAKGKVLQAYPERNRVLVEGVNRIKKHTAISTN
MUL_0805|M.ulcerans_Agy99 MKVHKGDTVLVVSGKDKGAKGKVLQAYPERNRVLVEGVNRIKKHTAISTN
MLBr_01848|M.leprae_Br4923 MKVHKGDTVLVIAGKDKGAKGKVLQAYPARNRVLVEGVNRIKKHTAISAN
MAV_4454|M.avium_104 MKVRKGDTVLVIAGKDKGAKGKVLKAYPDRERVLVEGVNRIKKHTAISTN
MAB_3806c|M.abscessus_ATCC_199 MKVHKGDTVLVIAGKDKGAKGKVIAAYPERSRVLVEGVNRIKKHTPQSAN
Mflv_5036|M.gilvum_PYR-GCK MKVRKGDTVLVISGKDKGAKGKVLVAYPDRNKVLVEGVNRIKKHTAESRT
Mvan_1333|M.vanbaalenii_PYR-1 MKVRKGDTVLVISGKDKGAKGKVLVAYPDRNKVLVEGVNRIKKHTPESRT
MSMEG_1466|M.smegmatis_MC2_155 MKVHKGDTVLVISGKDKGAKGKVLVAYPDRNKVLVEGVNRIKKHTAVSAN
TH_0312|M.thermoresistible__bu MKVRKGDTVLVISGKDKGAKGKVLQAYPDRQKVLVEGVNRIKKHTAVSRN
***:*******::**********: *** *.:*************. * .
Mb0736|M.bovis_AF2122/97 QRGARSGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRVDEETGKRVRISKR
Rv0715|M.tuberculosis_H37Rv QRGARSGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRVDEETGKRVRISKR
MMAR_1046|M.marinum_M QRGAKSGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRVDEETGKRVRISKR
MUL_0805|M.ulcerans_Agy99 QRGAKSGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRVDEETGKRVRISKR
MLBr_01848|M.leprae_Br4923 QRGAQAGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRVDEETDKRVRISKR
MAV_4454|M.avium_104 QRGAQSGGIVTQEAPIHVSNVMVVDSDGKPARVGYRLDEETGKRVRISKR
MAB_3806c|M.abscessus_ATCC_199 ERGASSGGIVTQEAPIHVSNVMVIDSDGQPTRIGYRVDEETGKKVRISRR
Mflv_5036|M.gilvum_PYR-GCK ERGAASGGIVTQEAPISVSNVMLLDSDGKPTRVGYRKDDETGKNVRIAKS
Mvan_1333|M.vanbaalenii_PYR-1 ERGASSGGIVTQEAPISVSNVMLLDSDGKPTRVGYRRDDETGKNVRIAKS
MSMEG_1466|M.smegmatis_MC2_155 ERGASSGGIVTQEAPIHVSNVMVVDSDGKPTRVGYRIDDETGKKVRIAKT
TH_0312|M.thermoresistible__bu ERGAQSGGIVTQEAPIHVSNVMVVDSDGKPTRIGYRFDEETGKKVRIAKT
:*** :********** *****::****:*:*:*** *:**.*.***::
Mb0736|M.bovis_AF2122/97 NGKDI--
Rv0715|M.tuberculosis_H37Rv NGKDI--
MMAR_1046|M.marinum_M NGKDIQA
MUL_0805|M.ulcerans_Agy99 NGKDIQA
MLBr_01848|M.leprae_Br4923 NGKDI--
MAV_4454|M.avium_104 NGKDI--
MAB_3806c|M.abscessus_ATCC_199 NGKDI--
Mflv_5036|M.gilvum_PYR-GCK NGKDL--
Mvan_1333|M.vanbaalenii_PYR-1 NGKDI--
MSMEG_1466|M.smegmatis_MC2_155 NGKDI--
TH_0312|M.thermoresistible__bu NGKDI--
****: