For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTELLGRDEFRAELENAIKGREAKNASFSKAWADGKLTKDHFARWAENHYHYVGPFADYLANIYSNTPDE FTGAKDFTLQNMYEEELADIRHTDLLIKFGEACGTTRERIEDPSNMNPITRGLQAWCYAVSQREHFVVAT AALVVGLESQVPSIYTKQIVPLREVYGFTEDEIEFFDLHITSDVVHGERGYQIVLDHADTPQLQQRCLQL VRWGAEMRFSYTKGLYDYYVAPDLEAAGV
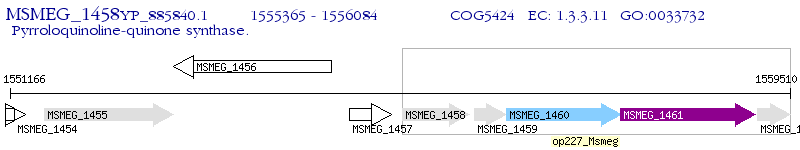
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1458 | - | - | 100% (239) | tena/thi-4 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1325 | - | 1e-130 | 92.05% (239) | TenA/THI-4 protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1458|M.smegmatis_MC2_155 MTELLGRDEFRAELENAIKGREAKNASFSKAWADGKLTKDHFARWAENHY
Mvan_1325|M.vanbaalenii_PYR-1 MTELLGRDEFRTELENAIKGREAKNASFSQAWAEGRLQRHHFARWAENHY
***********:*****************:***:*:* :.**********
MSMEG_1458|M.smegmatis_MC2_155 HYVGPFADYLANIYSNTPDEFTGAKDFTLQNMYEEELADIRHTDLLIKFG
Mvan_1325|M.vanbaalenii_PYR-1 HYVGPFADYLANIYSNTPDNFTGAKDFTLQNMYEEELADIRHTDLLIKFG
*******************:******************************
MSMEG_1458|M.smegmatis_MC2_155 EACGTTRERIEDPSNMNPITRGLQAWCYAVSQREHFVVATAALVVGLESQ
Mvan_1325|M.vanbaalenii_PYR-1 EACGTTKERIEDPDNMNAITRGLQSWCYAVSQREHFVVATAALVVGLESQ
******:******.***.******:*************************
MSMEG_1458|M.smegmatis_MC2_155 VPSIYTKQIVPLREVYGFTEDEIEFFDLHITSDVVHGERGYQIVLDHADT
Mvan_1325|M.vanbaalenii_PYR-1 VPSIYMKQIVPLREVYKFTEDEIEFFDLHITSDVVHGERGYQIVLDHADT
***** ********** *********************************
MSMEG_1458|M.smegmatis_MC2_155 PQLQQRCLQLVRWGAEMRFSYTKGLYDYYVAPDLEAAGV
Mvan_1325|M.vanbaalenii_PYR-1 PQLQQRCLQMVRWGAEMRFSYTKGLYDTYVAPELEPAAV
*********:***************** ****:**.*.*