For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DTPLTAAHWIYLAGIVVILLTMAVRKNIVVPAVTATLLTAWAFSGSLITGLSAIFNASLTAAKELFNIFL IIAIVTAMLGALREMGADRMMVTPFRRVMRAGTSSFIVLAIVTYVISLFFWPTPAVPLVGAVLIPVAIRA GLSPMSVGMVIAIAGQGMALSSDYIIKVAPGISAKAAGVDPDSVADKAMVLSLIVGLTALGITYVMQRRT WRNPSPELLVEWENAAERRGVGDDDEDSPSGGPAGSPSATAGSNGRKPAPAGPTVTDGPTRSDGDTAVAT VKAVKAAVIVEAIVDTGAETGEQEPKKLSAKVFALLVPLVYLAFIVYLLLGKFTNVVPPLQGGDAAAIVG GLAAMLLFAAAATNDKRNFLETSSQHVVDGLVFAFKAMGIVLPIAGFFFIGNGDFSAAILGMPADATGPA FLFDLITAAQSHLSPNPVVTSFAVLFVGLIAGLEGSGFSGLPLTGSLSGALGPVVHMDPSTLAAIGQMGN IWSGGGTLVAWSSLVAVAGFARVPVLSLARKCFLPVMAGLVLSTVVAIIIF*
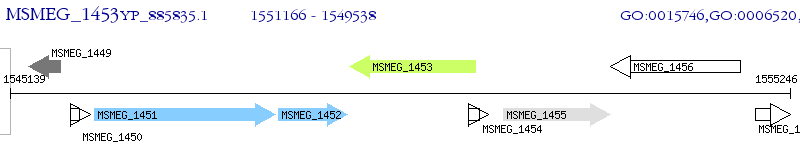
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1453 | - | - | 100% (542) | citrate transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1321 | - | 0.0 | 75.83% (542) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1453|M.smegmatis_MC2_155 MDTPLTAAHWIYLAGIVVILLTMAVRKNIVVPAVTATLLTAWAFSGSLIT
Mvan_1321|M.vanbaalenii_PYR-1 MDIHLSAAHWVYLAGIVVILVTMALRKNIVVPAVVATLLTAWVFSGSIVT
** *:****:*********:***:*********.*******.****::*
MSMEG_1453|M.smegmatis_MC2_155 GLSAIFNASLTAAKELFNIFLIIAIVTAMLGALREMGADRMMVTPFRRVM
Mvan_1321|M.vanbaalenii_PYR-1 GLSSIFNASLVAADELFNIFLIIALVTAMLGALREMGADRLMVTPFRRVM
***:******.**.**********:***************:*********
MSMEG_1453|M.smegmatis_MC2_155 RAGTSSFIVLAIVTYVISLFFWPTPAVPLVGAVLIPVAIRAGLSPMSVGM
Mvan_1321|M.vanbaalenii_PYR-1 RAGTSSFVVLAIVTYVISLFFWPTPAVPLVGAVLIPVAIRAGLSPMSVGM
*******:******************************************
MSMEG_1453|M.smegmatis_MC2_155 VIAIAGQGMALSSDYIIKVAPGISAKAAGVDPDSVADKAMVLSLIVGLTA
Mvan_1321|M.vanbaalenii_PYR-1 VVAIAGQGMALSSDYIIKVAPGISATAAGVDADGVADKSLVLSLIVGGVA
*:***********************.*****.*.****::******* .*
MSMEG_1453|M.smegmatis_MC2_155 LGITYVMQRRTWRNPSPELLVEWENAAERRGVGDDDEDSPSGGPAGSPSA
Mvan_1321|M.vanbaalenii_PYR-1 LGITYVTQRRTWRTPSPELLVAWENKADKLGAPDDD--TAAGDEFG----
****** ******.******* *** *:: *. *** :.:*. *
MSMEG_1453|M.smegmatis_MC2_155 TAGSNGRKPAPAGPTVTDGPTRSDGDTAVATVKAVKAAVIVEAIVDTGAE
Mvan_1321|M.vanbaalenii_PYR-1 -----GTRPAP--------------DPAPAPVAG--GAPTATMVLDTSSI
* :*** *.* *.* . .* . ::**.:
MSMEG_1453|M.smegmatis_MC2_155 TGEQEPKKLSAKVFALLVPLVYLAFIVYLLLGKFTNVVPPLQGGDAAAIV
Mvan_1321|M.vanbaalenii_PYR-1 EVADSPKTLSAKTFAVLVPTAYLVFIVYMLLGKFTTAVPPLKGGDAAGIV
:.**.****.**:*** .**.****:******..****:*****.**
MSMEG_1453|M.smegmatis_MC2_155 GGLAAMLLFAAAATNDKRNFLETSSQHVVDGLVFAFKAMGIVLPIAGFFF
Mvan_1321|M.vanbaalenii_PYR-1 GGIAAMLLFAACLTNDKRACLESSSRHIVDGLVFAFKAMGIVLPIAGFFF
**:********. ***** **:**:*:**********************
MSMEG_1453|M.smegmatis_MC2_155 IGNGDFSAAILGMPADATGPAFLFDLITAAQSHLSPNPVVTSFAVLFVGL
Mvan_1321|M.vanbaalenii_PYR-1 IGNGDFSARILGMPEDATGPALLFDLINAGLSHVAPNAFVVAFTVLFVGL
******** ***** ******:*****.*. **::**..*.:*:******
MSMEG_1453|M.smegmatis_MC2_155 IAGLEGSGFSGLPLTGSLSGALGPVVHMDPSTLAAIGQMGNIWSGGGTLV
Mvan_1321|M.vanbaalenii_PYR-1 IAGLEGSGFSGLPLTGSLSGALGSAAGMDPSTLAAIGQMGNIWSGGGTLV
***********************... ***********************
MSMEG_1453|M.smegmatis_MC2_155 AWSSLVAVAGFARVPVLSLARKCFLPVMAGLVLSTVVAIIIF
Mvan_1321|M.vanbaalenii_PYR-1 AWSSLVAVAGFARVPVLTLARKCFLPVMAGLVLSTLFAIIAF
*****************:*****************:.*** *