For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGKGGRNAHGRITTRHKGGGHKRAYRVIDFRRH DKDGVNAKVAHIEYDPNRTANIALLHYLDGEKRYIIAPQGLKQGDVIESGANADIKPGNNLPLRNIPAGT VIHAVELRPGGGAKLARSAGVSIQLLGKEGTYAALRMPSGEIRRVDVRCRATVGEVGNAEQSNINWGKAG RMRWKGKRPTVRGVVMNPVDHPHGGGEGKTSGGRHPVSPWGKPEGRTRKPNKPSDKLIVRRRRTGKKR
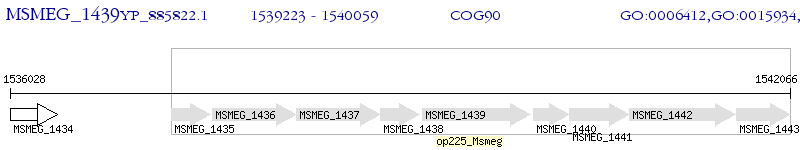
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1439 | rplB | - | 100% (278) | 50S ribosomal protein L2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0724 | rplB | 1e-153 | 92.78% (277) | 50S ribosomal protein L2 |
| M. gilvum PYR-GCK | Mflv_5046 | rplB | 1e-157 | 93.88% (278) | 50S ribosomal protein L2 |
| M. tuberculosis H37Rv | Rv0704 | rplB | 1e-153 | 92.78% (277) | 50S ribosomal protein L2 |
| M. leprae Br4923 | MLBr_01860 | rplB | 1e-150 | 89.89% (277) | 50S ribosomal protein L2 |
| M. abscessus ATCC 19977 | MAB_3817c | rplB | 1e-155 | 92.45% (278) | 50S ribosomal protein L2 |
| M. marinum M | MMAR_1034 | rplB | 1e-154 | 93.50% (277) | 50S ribosomal protein L2, RplB |
| M. avium 104 | MAV_4468 | rplB | 1e-155 | 93.50% (277) | 50S ribosomal protein L2 |
| M. thermoresistible (build 8) | TH_0404 | rplB | 1e-157 | 94.96% (278) | PROBABLE 50S ribosomal protein L2 RPLB |
| M. ulcerans Agy99 | MUL_0793 | rplB | 1e-154 | 93.14% (277) | 50S ribosomal protein L2 |
| M. vanbaalenii PYR-1 | Mvan_1308 | rplB | 1e-157 | 93.88% (278) | 50S ribosomal protein L2 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01860|M.leprae_Br4923 MAIRKYKPTTSGRRGASVSDFTDITRTKPEKALMRSLHGHGGRNVHGRIT
MAV_4468|M.avium_104 MAIRKYKPTSPGRRGASVSDFSEVTRSTPEKSLVRPLHGHGGRNAHGRIT
Mb0724|M.bovis_AF2122/97 MAIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGRGGRNAHGRIT
Rv0704|M.tuberculosis_H37Rv MAIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGRGGRNAHGRIT
MMAR_1034|M.marinum_M MAIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGHGGRNAHGRIT
MUL_0793|M.ulcerans_Agy99 MAIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGHGGRNAHGRIT
MAB_3817c|M.abscessus_ATCC_199 MAIRKYKPTTPGRRGSSVADFSEITRSTPEKSLIRPLHGTGGRNAHGRIT
MSMEG_1439|M.smegmatis_MC2_155 MGIRKYKPTTPGRRGASVSDFAEITRSTPEKSLVRPLHGKGGRNAHGRIT
TH_0404|M.thermoresistible__bu MAIRKYKPTSPGRRGASVSDFAEITRSHPEKSLVRPLHGKGGRNAHGRIT
Mflv_5046|M.gilvum_PYR-GCK MAIRKYKPTTPGRRGSSVSDFAEITRDHPEKSLIRPLHGKGGRNAHGRIT
Mvan_1308|M.vanbaalenii_PYR-1 MAIRKYKPTTPGRRGSSVSDFAEITRDHPEKSLVRPLHGRGGRNAHGRIT
*.*******:.****:**:**:::** ***:*:*.*** ****.*****
MLBr_01860|M.leprae_Br4923 TRHKGGGHKRAYRLIDFRRNDTDGVNAKVAHIEYDPNRTANIALLHFLDG
MAV_4468|M.avium_104 TRHKGGGHKRAYRVIDFRRNDKDGVNAKVAHIEYDPNRTANIALLHFLDG
Mb0724|M.bovis_AF2122/97 TRHKGGGHKRAYRMIDFRRNDKDGVNAKVAHIEYDPNRTARIALLHYLDG
Rv0704|M.tuberculosis_H37Rv TRHKGGGHKRAYRMIDFRRNDKDGVNAKVAHIEYDPNRTARIALLHYLDG
MMAR_1034|M.marinum_M TRHKGGGHKRAYRVIDFRRNDKDGVNAKVAHIEYDPNRTARIALLHYLDG
MUL_0793|M.ulcerans_Agy99 TRHKGGGHKRAYRVIDFGRNDKDGVNAKVAHIEYDPNRTARIALLHYLDG
MAB_3817c|M.abscessus_ATCC_199 TRHKGGGHKRAYRLIDFRRHDKDGVNAKVAHIEYDPNRTARIALLHFLDG
MSMEG_1439|M.smegmatis_MC2_155 TRHKGGGHKRAYRVIDFRRHDKDGVNAKVAHIEYDPNRTANIALLHYLDG
TH_0404|M.thermoresistible__bu TRHKGGGHKRAYRVIDFRRHDKDGVNAKVAHIEYDPNRTAHIALLHYLDG
Mflv_5046|M.gilvum_PYR-GCK TRHKGGGHKRAYRVIDFRRHDKDGVDAKVAHIEYDPNRTANIALLHYLDG
Mvan_1308|M.vanbaalenii_PYR-1 TRHKGGGHKRAYRVIDFRRHDKDGVNAKVAHIEYDPNRTANIALLHYLDG
*************:*** *:*.***:**************.*****:***
MLBr_01860|M.leprae_Br4923 KKRYILAPQGLSQGDVVESGANADIKPGNNLPLRNIPAGTLIHAVELRPG
MAV_4468|M.avium_104 EKRYIIAPQGLSQGDVVESGPNADIKPGNNLPLRNIPAGTVIHAVELRPG
Mb0724|M.bovis_AF2122/97 EKRYIIAPNGLSQGDVVESGANADIKPGNNLPLRNIPAGTLIHAVELRPG
Rv0704|M.tuberculosis_H37Rv EKRYIIAPNGLSQGDVVESGANADIKPGNNLPLRNIPAGTLIHAVELRPG
MMAR_1034|M.marinum_M EKRYIIAPNGLSQGDVVESGANADIKPGNNLPLRNIPAGTLVHAVELRPG
MUL_0793|M.ulcerans_Agy99 EKRYIIAPNGLSQGDVVESGANADIKPGNNLPLRNIPAGTLVHAVELRPG
MAB_3817c|M.abscessus_ATCC_199 EKRYIIAPVGLSQGDVVESGANADIKPGNNLPLRNIPTGTTVHAVELRPG
MSMEG_1439|M.smegmatis_MC2_155 EKRYIIAPQGLKQGDVIESGANADIKPGNNLPLRNIPAGTVIHAVELRPG
TH_0404|M.thermoresistible__bu EKRYIIAPQGLRQGDVVESGANADIKPGNNLPLRNIPAGTVIHAVELRPG
Mflv_5046|M.gilvum_PYR-GCK EKRYIIAPQGLKQGAIVESGANADIKPGNNLPLRNIPAGTLIHAVELRPG
Mvan_1308|M.vanbaalenii_PYR-1 EKRYIIAPQGLKQGAIVESGANADIKPGNNLPLRNIPAGTLVHAVELRPG
:****:** ** ** ::***.****************:** :********
MLBr_01860|M.leprae_Br4923 GGAKLARSAGSSIQLLGKESSYASLRMPSGEIRRVDVRCRATVGEVGNAE
MAV_4468|M.avium_104 GGAKLARSAGSSIQLLGKEGSYASLRMPSGEIRRVDVRCRATVGEVGNAE
Mb0724|M.bovis_AF2122/97 GGAKLARSAGSSIQLLGKEASYASLRMPSGEIRRVDVRCRATVGEVGNAE
Rv0704|M.tuberculosis_H37Rv GGAKLARSAGSSIQLLGKEASYASLRMPSGEIRRVDVRCRATVGEVGNAE
MMAR_1034|M.marinum_M GGAKLARSAGSSIQLLGKEASYASLRMPSGEIRRVDVRCRATVGEVGNAE
MUL_0793|M.ulcerans_Agy99 GGAKLARSAGSSIQLLGKEASYASLRMPSGEIRRVDVRCRATVGEVGNAE
MAB_3817c|M.abscessus_ATCC_199 GGAKLARSAGSGIQLLGKEGAYASLRMPSGEIRRVDVRCRATVGEVGNAE
MSMEG_1439|M.smegmatis_MC2_155 GGAKLARSAGVSIQLLGKEGTYAALRMPSGEIRRVDVRCRATVGEVGNAE
TH_0404|M.thermoresistible__bu GGAKLARSAGASIQLLGKEGSYASLRMPSGEIRRVDVRCRATVGEVGNAE
Mflv_5046|M.gilvum_PYR-GCK GGAKMARSAGASIQLLGKEGTYASLRMPSGEIRRVDVRCRATVGEVGNAE
Mvan_1308|M.vanbaalenii_PYR-1 GGAKMARSAGASIQLLGKEGSYASLRMPSGEIRRVDVRCRATVGEVGNAE
****:***** .*******.:**:**************************
MLBr_01860|M.leprae_Br4923 QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
MAV_4468|M.avium_104 QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
Mb0724|M.bovis_AF2122/97 QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
Rv0704|M.tuberculosis_H37Rv QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
MMAR_1034|M.marinum_M QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
MUL_0793|M.ulcerans_Agy99 QANINWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
MAB_3817c|M.abscessus_ATCC_199 QANINWGKAGRMRWKGKRPTVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
MSMEG_1439|M.smegmatis_MC2_155 QSNINWGKAGRMRWKGKRPTVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
TH_0404|M.thermoresistible__bu QINISWGKAGRMRWKGKRPSVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
Mflv_5046|M.gilvum_PYR-GCK QANINWGKAGRMRWKGKRPTVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
Mvan_1308|M.vanbaalenii_PYR-1 QANINWGKAGRMRWKGKRPTVRGVVMNPVDHPHGGGEGKTSGGRHPVSPW
* **.**************:******************************
MLBr_01860|M.leprae_Br4923 GKPEGRTRKPNKSSNKLIVRRRRTGKKHAR
MAV_4468|M.avium_104 GKPEGRTRHPNKASNKLIVRRRRTGKKHGR
Mb0724|M.bovis_AF2122/97 GKPEGRTRNANKSSNKFIVRRRRTGKKHSR
Rv0704|M.tuberculosis_H37Rv GKPEGRTRNANKSSNKFIVRRRRTGKKHSR
MMAR_1034|M.marinum_M GKPEGRTRNPNKASNKLIVRRRRTGKKHGR
MUL_0793|M.ulcerans_Agy99 GKPEGRTRNPNKASNKLIVRRRRTGKKHGR
MAB_3817c|M.abscessus_ATCC_199 GKPEGRTRKPNKASDKLIVRRRRTGKKR--
MSMEG_1439|M.smegmatis_MC2_155 GKPEGRTRKPNKPSDKLIVRRRRTGKKR--
TH_0404|M.thermoresistible__bu GKLEGRTRRPNKPSDKLIVRRRRTGKKR--
Mflv_5046|M.gilvum_PYR-GCK GKPEGRTRKPNKASDKLIVRRRRTGKNKR-
Mvan_1308|M.vanbaalenii_PYR-1 GKPEGRTRKPKKASDKLIVRRRRTGKKR--
** *****..:*.*:*:*********::