For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVPDVRRMTARSVVLSVMLGAHPAWASSAELIRLTSDFGIREQTLRVALTRMVSAGDLVRSADGYRLSD RLMVRQRRQDDAIDPKVHDHDGEWLTVVVTSVGTDARTRAWMRNTLQQYRFGELREGVWLRPDNLDQALP GEITDRVRVLRTRDDNPRDLAARLWDLPGWARTGELLLEAMADATDVPARFVVAAGIVRHLLVDPVLPDD LLPASWPGAALRAAYNDFAAELVARRDRTELMEAT
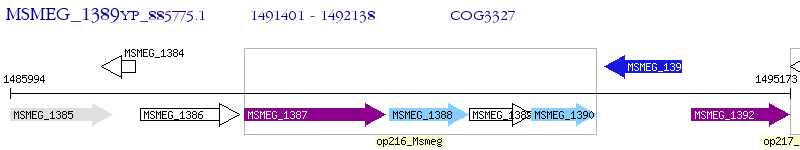
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1389 | - | - | 100% (245) | hypothetical protein MSMEG_1389 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0693 | - | 2e-89 | 66.24% (237) | hypothetical protein Mb0693 |
| M. gilvum PYR-GCK | Mflv_5085 | - | 3e-94 | 70.46% (237) | PaaX domain-containing protein, C- domain |
| M. tuberculosis H37Rv | Rv0674 | - | 3e-89 | 66.24% (237) | hypothetical protein Rv0674 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3858c | - | 2e-73 | 57.68% (241) | hypothetical protein MAB_3858c |
| M. marinum M | MMAR_1003 | - | 3e-87 | 64.88% (242) | putative regulatory protein |
| M. avium 104 | MAV_4498 | - | 4e-96 | 72.93% (229) | hypothetical protein MAV_4498 |
| M. thermoresistible (build 8) | TH_1897 | - | 1e-12 | 73.17% (41) | glyoxalase family protein |
| M. ulcerans Agy99 | MUL_0755 | - | 1e-87 | 64.88% (242) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1271 | - | 1e-99 | 75.00% (236) | PaaX domain-containing protein, C- domain |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5085|M.gilvum_PYR-GCK ---------------------MRRPAN-RMTARSVVLSVLLGAHP-----
Mvan_1271|M.vanbaalenii_PYR-1 -----------------------------MTARSVVLSVLLGAHP-----
MSMEG_1389|M.smegmatis_MC2_155 ---------------------MTVPDVRRMTARSVVLSVMLGAHP-----
Mb0693|M.bovis_AF2122/97 --------------------------MPAMTARSVVLSVLLGAHP-----
Rv0674|M.tuberculosis_H37Rv --------------------------MPAMTARSVVLSVLLGAHP-----
MMAR_1003|M.marinum_M --------------------------MAAMTARSVVLSVLLGAHP-----
MUL_0755|M.ulcerans_Agy99 --------------------------MAAMTARSVVLSVLLGAHP-----
MAV_4498|M.avium_104 --------------------------MPDLTARSVVLSVLLGAHP-----
MAB_3858c|M.abscessus_ATCC_199 --------------------MDNPVDHHPMTARSVILSLLLGAHP-----
TH_1897|M.thermoresistible__bu MNQHHSRDPLDVLRGGDRPVQPDPAFAARLRARLESAARFLADRPHTEGV
: ** : :*. :*
Mflv_5085|M.gilvum_PYR-GCK -AWATAAELIRLTSDFGIRESTV---------------------------
Mvan_1271|M.vanbaalenii_PYR-1 -AWATAAELIRLTSDFGIRESTV---------------------------
MSMEG_1389|M.smegmatis_MC2_155 -AWASSAELIRLTSDFGIREQTL---------------------------
Mb0693|M.bovis_AF2122/97 -AWATASELIQLTADFGIKETTL---------------------------
Rv0674|M.tuberculosis_H37Rv -AWATASELIQLTADFGIKETTL---------------------------
MMAR_1003|M.marinum_M -AWASASELIRLTADFGIKETAL---------------------------
MUL_0755|M.ulcerans_Agy99 -AWASASELIRLTADFGIKETAL---------------------------
MAV_4498|M.avium_104 -AHARASELVRLTSDFGIKESTL---------------------------
MAB_3858c|M.abscessus_ATCC_199 -AELSVREIRALTTLFGISDTTV---------------------------
TH_1897|M.thermoresistible__bu VMSGTQTALTELTGSTGTAEPAATTPPRPAAVPYLNVADARAAIDWYVEA
: ** * : :
Mflv_5085|M.gilvum_PYR-GCK ---------------RVALTRMVSAGDLVRSEDGYRLS------------
Mvan_1271|M.vanbaalenii_PYR-1 ---------------RVALTRMVGAGDLVRSEDGYRLS------------
MSMEG_1389|M.smegmatis_MC2_155 ---------------RVALTRMVSAGDLVRSADGYRLS------------
Mb0693|M.bovis_AF2122/97 ---------------RVALTRMVGAGDLVRSADGYRLS------------
Rv0674|M.tuberculosis_H37Rv ---------------RVALTRMVGAGDLVRSADGYRLS------------
MMAR_1003|M.marinum_M ---------------RVALTRMVSGGDLIRSADGYRLS------------
MUL_0755|M.ulcerans_Agy99 ---------------RVALTRMVSGGDLIRSADGYRLS------------
MAV_4498|M.avium_104 ---------------RVALTRMVNAGDLIRSADGYRLS------------
MAB_3858c|M.abscessus_ATCC_199 ---------------RVALTRMVAAGDLVRTEAGYRLA------------
TH_1897|M.thermoresistible__bu LGATLSGEPVVMDDGRIGHAELTLAGGVFYLADEYPELGLRAPAPEAVSM
*:. :.:. .*.:. *
Mflv_5085|M.gilvum_PYR-GCK ---------GRLLARQRRQDDVLEPRRGEWDGLWTTLVITSVGADARTRA
Mvan_1271|M.vanbaalenii_PYR-1 ---------DRLLARQRRQDDAITPRRHAWDGSWTMLVITSVGTDARTRA
MSMEG_1389|M.smegmatis_MC2_155 ---------DRLMVRQRRQDDAIDPKVHDHDGEWLTVVVTSVGTDARTRA
Mb0693|M.bovis_AF2122/97 ---------DRLLARQRRQDEAMRPRTRAWHGNWHMLIVTSIGTDARTRA
Rv0674|M.tuberculosis_H37Rv ---------DRLLARQRRQDEAMRPRTRAWHGNWHMLIVTSIGTDARTRA
MMAR_1003|M.marinum_M ---------DRLLARQARQDEAMRPRVRPWQGNWHLVIVTSVGTDARTRA
MUL_0755|M.ulcerans_Agy99 ---------DRLLARQARQDEAMRPRVRPWQGNWHLVIVTSVGTDARTRA
MAV_4498|M.avium_104 ---------DRLLARQRRQDDAIDPKVRPWRGQWVTLIVTSVGDDARTRA
MAB_3858c|M.abscessus_ATCC_199 ---------ERLQARQLRQDEACDPHRGSWDGTWKQLVITSVGRDARDRN
TH_1897|M.thermoresistible__bu SLVLPVADTDAALQRARRHGATVQREPYEAHGARSAAIVDPFGHRWMLSG
* *:. . . * :: ..*
Mflv_5085|M.gilvum_PYR-GCK HLRNTLQNHRFAELR-DGVWMRPANVEVQLGANITEKARILQARDERPAE
Mvan_1271|M.vanbaalenii_PYR-1 QLRNTLQDNRFAELR-EGVWMRPANVDLELGPDITEKVRILAARDDEPAE
MSMEG_1389|M.smegmatis_MC2_155 WMRNTLQQYRFGELR-EGVWLRPDNLDQALPGEITDRVRVLRTRDDNPRD
Mb0693|M.bovis_AF2122/97 ALRTCMHHKRFGELR-EGVWMRPDNLDLDLESDVAARVRMLTARDEAPAD
Rv0674|M.tuberculosis_H37Rv ALRTCMHHKRFGELR-EGVWMRPDNLDLDLESDVAARVRMLTARDEAPAD
MMAR_1003|M.marinum_M ALRTTMHDKRFAELR-EGVWLRPDNLDLELAADVAERVRVLTARDDAPAE
MUL_0755|M.ulcerans_Agy99 ALRTTMHDKRFAELR-EGVWLRPDNLDLELAADVAERVRVLTARDDAPAE
MAV_4498|M.avium_104 ALRNAMHDKRFGELR-EGVWMRPDNIDTQLGPDVTDRARVITARDDDPAD
MAB_3858c|M.abscessus_ATCC_199 DLRTTLRQSRFGEFR-EGVWLRPENLEVDLPPSVTDHVWILHTRTDRPEV
TH_1897|M.thermoresistible__bu PLTGASARIQHGDIGYVALWVPDPQRAAQFYRQVLGWQWDPDSNQVTNTT
: :..:: .:*: : : .: :.
Mflv_5085|M.gilvum_PYR-GCK LATHLWDLTGWAVDGRRLLK---EMDDAHDVPTRFVAAATIVRHLLTDPV
Mvan_1271|M.vanbaalenii_PYR-1 LAARLWDLPGWAATGRELLT---RIADADDVPGRFVVAAAMVRHLLTDPV
MSMEG_1389|M.smegmatis_MC2_155 LAARLWDLPGWARTGELLLE---AMADATDVPARFVVAAGIVRHLLVDPV
Mb0693|M.bovis_AF2122/97 LAGQLWDLSGWTEAGHRLLG---DMAAATDMPGRFVVAAAMVRHLLTDPM
Rv0674|M.tuberculosis_H37Rv LAGQLWDLSGWTEAGHRLLG---DMAAATDMPGRFVVAAAMVRHLLTDPM
MMAR_1003|M.marinum_M LAGQLWDLTEWAGVGDRLLGEMAEMAEAADIPGQFALAAAMVRHLLSDPM
MUL_0755|M.ulcerans_Agy99 LAGQLWDLTGWAGVGDRLLGEMAEMAEAAGIPGQFALAAAMVRHLLSDPM
MAV_4498|M.avium_104 LAGRLWDLPGWARAGHRLLD---EMAAAPDVPGRFVVAAAMVRHLLTDPV
MAB_3858c|M.abscessus_ATCC_199 LAAQLWDLTGWSTYATALLR---WWDQAEAIPARFVLAAAMVRHLLADPV
TH_1897|M.thermoresistible__bu GPIGIHRVPDARTMFCCYAVTDLDAARQAIIDGGGPPXXXMVRHLLADPV
. : :. : :***** **:
Mflv_5085|M.gilvum_PYR-GCK LPDELLPADWPGADLRAAYNDFAAEMVARRDDVKLTEAG-
Mvan_1271|M.vanbaalenii_PYR-1 LPDELLATDWPGAALRSAYNDFAAEMVARRDGVELMEAR-
MSMEG_1389|M.smegmatis_MC2_155 LPDDLLPASWPGAALRAAYNDFAAELVARRDRTELMEAT-
Mb0693|M.bovis_AF2122/97 LPAELLPADWPGAGLRAAYHDFATAMAKRRDATQLLEVT-
Rv0674|M.tuberculosis_H37Rv LPAELLPADWPGAGLRAAYHDFATAMAKRRDATQLLEVT-
MMAR_1003|M.marinum_M LPEELLPAKWPGAQLRSAYHDFATELAKRRGTNQLLEAK-
MUL_0755|M.ulcerans_Agy99 LPEELLPAKWPGAQLRSAYHDFATELAKRRGTNQLLEAK-
MAV_4498|M.avium_104 LPDELLPPDWPGTRLRAAYHDFAAELMARRDP-LFAEAT-
MAB_3858c|M.abscessus_ATCC_199 LPDELLPADWPADEMRARYREFKDELVTLRDTAEMEEASL
TH_1897|M.thermoresistible__bu LPDELLPAGWPGGAVRDAYRVFAAEMAARRDAGLVEAFD-
** :**.. **. :* *. * : *. .