For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTMWATPGDIAAKVRRRWDDGSLLRAYGEGEPFAPIEVPLRGPTPAQIGDDVARVRDWVTALDAGRHDD ARYMLEWKSVGGKKFGRNMLPARAVICSYGQAWTLLGVTAAVGRFDELLASAHDHPRIRQWVIANPHRAL ALAADMPKLIAACEWLDRHRHSQRYLREISAPGVDTKFVERRRSDLAGILGVPSSAQGFLTELGLRSKPS LVRLRPAPSLGLPAPLSELAVRPDELAQLDLRPRTAVIVENEISYLSVDVPEAGVVIWGKGFDVDNVGRL PWLAGVRVVYWGDIDTHGFAILDRLRAWQPTAESVLMDRATLIAHRDRWVVEDRPARSILPRLSPEEQQL YRELVEDSLGDRVRLEQERIDWGWVEGRLKAIYQGRIDS
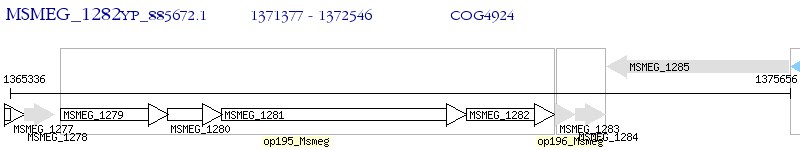
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1282 | - | - | 100% (389) | putative cytoplasmic protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1134 | - | 1e-159 | 70.93% (375) | hypothetical protein Mvan_1134 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1282|M.smegmatis_MC2_155 MST-MWATPGDIAAKVRRRWDDGSLLRAYGEGEPFAPIEVPLRGPTPAQI
Mvan_1134|M.vanbaalenii_PYR-1 MSARGWTTVGDIVARLRRRWDDGSLLTGHAKAEPFEPVEVPLRGPASAAI
**: *:* ***.*::********** .:.:.*** *:*******:.* *
MSMEG_1282|M.smegmatis_MC2_155 GDDVARVRDWVTALDAGRHDDARYMLEWKSVGGKKFGRNMLPARAVICSY
Mvan_1134|M.vanbaalenii_PYR-1 GDDIAAVRDWVAALDAGRRDDTCYELEWKSVGGRKIGRNELPARAVLRRR
***:* *****:******:**: * ********:*:*** ******:
MSMEG_1282|M.smegmatis_MC2_155 GQAWTLLGVTAAVGRFDELLASAHDHPRIRQWVIANPHRALALAADMPKL
Mvan_1134|M.vanbaalenii_PYR-1 GQVWALLGVTGTVRRFDGLLEVSRRQPRVQEWITEHPHRALALASQIPQL
**.*:*****.:* *** ** :: :**:::*: :********:::*:*
MSMEG_1282|M.smegmatis_MC2_155 IAACEWLDRHRHSQRYLREISAPGVDTKFVERRRSDLAGILGVPSSAQGF
Mvan_1134|M.vanbaalenii_PYR-1 IAAYEWLDAHRHSRRYLREISAPGVDTKFVERHRTELAGMLGVAASSSGF
*** **** ****:******************:*::***:***.:*:.**
MSMEG_1282|M.smegmatis_MC2_155 LTELGLRSKPSLVRLRPAPSLGLPAPLSELAVRPDELAQLDLRPRTAVIV
Mvan_1134|M.vanbaalenii_PYR-1 ISDLGLRHKPALVRLRPSPSLGLPAALTELAVRAGELARLHVQPRVAVIV
:::**** **:******:*******.*:*****..***:*.::**.****
MSMEG_1282|M.smegmatis_MC2_155 ENEISYLSVDVPEAGVVIWGKGFDVDNVGRLPWLAGVRVVYWGDIDTHGF
Mvan_1134|M.vanbaalenii_PYR-1 ENEISYLSVDVPDDGIVLWGRGFDVDNVGRLPWLGGAEVVYWGDIDTHGF
************: *:*:**:*************.*..************
MSMEG_1282|M.smegmatis_MC2_155 AILDRLRAWQPTAESVLMDRATLIAHRDRWVVEDRPARSILPRLSPEEQQ
Mvan_1134|M.vanbaalenii_PYR-1 AILDRLRAWLPETRSVLMDRETLLAHHDRWVSEDRPARSALTRLTVDEFG
********* * :.****** **:**:**** ******* *.**: :*
MSMEG_1282|M.smegmatis_MC2_155 LYRELVEDSLGDRVRLEQERIDWGWVEGRLKAIYQGRIDS
Mvan_1134|M.vanbaalenii_PYR-1 LYTDLVEAALGDRVRLEQERIDWNWVERHLSLAT------
** :*** :**************.*** :*.