For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTRGPQRLAADHSRVLLRLFVPGQEGFEHQESRAGKVLKRILALDDNEVRATLAGVVAGYGERHRDLISV FRHHAGELADRLDPGRQLSEARMLLIGAMFTNEYAIEGAALCNPSMVAHPDQSGVPRGCLRFVMSVRGIG EGHCSSIGFRTGVVDSAGSVVVDDAVPFTATGSTTPPVLDAAVFRGYLAQLRHTGEIADYVLNALGDRFS RSDLDARLAELHRNRSTRQQAAETIDAIRAVADRAYGVVFPEDTALSERVLWPATSAEAAGMEDARFVRF VDDDGSITFYASYTAYSGNHISQQLLATTDFRTFTSVPMVGPAAANKGLALFPRRIGGRYAAMSRSDRES NTVAFTDHLAVWTDAAPCQQPVAPWEALQLGNCGPPIETDAGWLVLTHGVGPMRTYRIGAILLDLQDPTR IVGRLTEPLLSPTEDERDGYVPNVVYSCGGLVHAGMLVLPYGIGDAAIGIATVPLPDLLCALRG
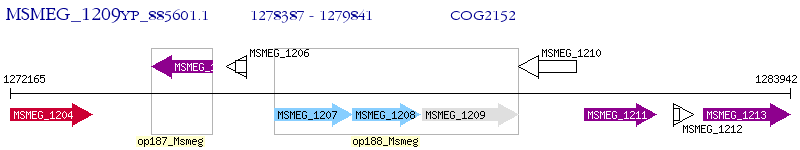
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1209 | - | - | 100% (484) | glycosidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5214 | - | 0.0 | 70.50% (478) | glycosidase, PH1107-related |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1054 | - | 0.0 | 71.90% (484) | glycosidase, PH1107-related |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5214|M.gilvum_PYR-GCK MTLAGERLATRSPQRVAADPSRVVTRLFVPGQEGFEAQDSRAGAVLSRIL
Mvan_1054|M.vanbaalenii_PYR-1 MTSTGIQLVTRSPQRVAADPGRVVTRLFVPGHEGFEHQESRAGAVLSRIL
MSMEG_1209|M.smegmatis_MC2_155 --------MTRGPQRLAADHSRVLLRLFVPGQEGFEHQESRAGKVLKRIL
**.***:*** .**: ******:**** *:**** **.***
Mflv_5214|M.gilvum_PYR-GCK SLTDDEVRATLHDVLSRFDGRHRDLKATLRRHAGEFADRLDKDCELDESR
Mvan_1054|M.vanbaalenii_PYR-1 ALTDDEVRAALQDVLIRFDGRHRDLTGTFRRHARELADRLDPTREFTEAR
MSMEG_1209|M.smegmatis_MC2_155 ALDDNEVRATLAGVVAGYGERHRDLISVFRHHAGELADRLDPGRQLSEAR
:* *:****:* .*: :. ***** ..:*:** *:***** :: *:*
Mflv_5214|M.gilvum_PYR-GCK MLLVGATFTNEYAIEGAALCNPSMVAHPDQSDVAAGSLRFVMSVRGIGEG
Mvan_1054|M.vanbaalenii_PYR-1 VLLLGATFTNEYSIEGAALCNPSVVAHPDQSGTVPGSLRFVMSVRGIGEG
MSMEG_1209|M.smegmatis_MC2_155 MLLIGAMFTNEYAIEGAALCNPSMVAHPDQSGVPRGCLRFVMSVRGIGEG
:**:** *****:**********:*******.. *.*************
Mflv_5214|M.gilvum_PYR-GCK HRSTIGFRSGVVDSAGNATVDEAARFAGTGRMRPTMLEAAVFRTELREQG
Mvan_1054|M.vanbaalenii_PYR-1 HRSSIGFRTGVVDSTGHATIDEPAPFASTGRVEPTLLDAAVFRTELRDKG
MSMEG_1209|M.smegmatis_MC2_155 HCSSIGFRTGVVDSAGSVVVDDAVPFTATGSTTPPVLDAAVFRGYLAQLR
* *:****:*****:* ..:*:.. *:.** *.:*:***** * :
Mflv_5214|M.gilvum_PYR-GCK RGGEAADYVFDALGELFTWSDLDEQLDKLHDHLRTRGHVEETITVIRSIA
Mvan_1054|M.vanbaalenii_PYR-1 CGGEAADYVFDALGALFTRSDLDERLERLRAHLSTRGHVEDTIATIRGVA
MSMEG_1209|M.smegmatis_MC2_155 HTGEIADYVLNALGDRFSRSDLDARLAELHRNRSTRQQAAETIDAIRAVA
** ****::*** *: **** :* .*: : ** :. :** .**.:*
Mflv_5214|M.gilvum_PYR-GCK ARAYAVEFPSDINLSECVLWPAMDAEHAGMEDARFVRFVDD-GSVSYHAT
Mvan_1054|M.vanbaalenii_PYR-1 ARCYAVEFPDDTTLSERVLWPEMEAEHAGMEDARFVRFVDDDGSIRYHAT
MSMEG_1209|M.smegmatis_MC2_155 DRAYGVVFPEDTALSERVLWPATSAEAAGMEDARFVRFVDDDGSITFYAS
*.*.* **.* *** **** .** ************** **: ::*:
Mflv_5214|M.gilvum_PYR-GCK YTAYSGSHISQQLLTTTDFRTFTSGPLVGRAAANKGLALFPRRINGRYAA
Mvan_1054|M.vanbaalenii_PYR-1 YTAYSGSHISQQLLTTADFQTFTSGPLVGSAAANKGLALFPRRIGGRYAA
MSMEG_1209|M.smegmatis_MC2_155 YTAYSGNHISQQLLATTDFRTFTSVPMVGPAAANKGLALFPRRIGGRYAA
******.*******:*:**:**** *:** **************.*****
Mflv_5214|M.gilvum_PYR-GCK MSRSDRETNTVAFADHLSVWPTASPCQAPAEVWETLQLGNCGPPIETGEG
Mvan_1054|M.vanbaalenii_PYR-1 MSRSDRETNTVAFADDLSVWTTALPCQQPAEVWETLQLGNCGPPIETDRG
MSMEG_1209|M.smegmatis_MC2_155 MSRSDRESNTVAFTDHLAVWTDAAPCQQPVAPWEALQLGNCGPPIETDAG
*******:*****:*.*:**. * *** *. **:************. *
Mflv_5214|M.gilvum_PYR-GCK WLVLTHGVGPMRTYSIGAILLDLDDPTRVIGRLRRPLLTPEPHERNGYVP
Mvan_1054|M.vanbaalenii_PYR-1 WLLLTHGVGPMRTYSIGAILLDLDDPTRVIGRLRRPLLTPAADDRDGYVP
MSMEG_1209|M.smegmatis_MC2_155 WLVLTHGVGPMRTYRIGAILLDLQDPTRIVGRLTEPLLSPTEDERDGYVP
**:*********** ********:****::*** .***:* .:*:****
Mflv_5214|M.gilvum_PYR-GCK NVVYSCGALVHAGTLVIPIGICDSAIGVATVPLADLMGELGR----
Mvan_1054|M.vanbaalenii_PYR-1 NVVYSCGALVHADTLVIPYGICDSAIGLATVPLPDLLAELAGSPRH
MSMEG_1209|M.smegmatis_MC2_155 NVVYSCGGLVHAGMLVLPYGIGDAAIGIATVPLPDLLCALRG----
*******.****. **:* ** *:***:*****.**: *