For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTSTTATVVGKLSTLDRFLPVWIGAAMAAGLLLGRWIPGLNTALESIQVDGISLPIALGLLIMMYPVLA KVRYDRLDTVTGDRKLLASSLILNWALGPALMFTLAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLAC GDREAAAVLVALNSIFQVVMFAVLGWFYLSVLPGWLGLEQATITTSPWQIARSVLIFLGIPLAAGYLSRR IGEKARGREWYESRFLPRVGPWALYGLLFTIVVLFALQGEQITSRPLDVVRIALPLLVYFAIMWGGGYLL GAVLGLSYERTTTLAFTAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVALVYVSLALRKRFN RAKEHTS*
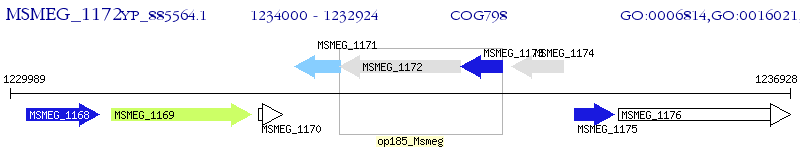
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1172 | arsB | - | 100% (358) | arsenical-resistance protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2676 | arsC | 1e-177 | 89.11% (349) | arsenic-transport integral membrane protein ArsC |
| M. gilvum PYR-GCK | Mflv_3469 | - | 1e-173 | 85.88% (347) | arsenical-resistance protein |
| M. tuberculosis H37Rv | Rv2643 | arsC | 1e-177 | 89.11% (349) | arsenic-transport integral membrane protein ArsC |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4863 | - | 1e-177 | 86.15% (361) | arsenic-transport integral membrane protein ArsC |
| M. marinum M | MMAR_2055 | arsC | 1e-176 | 89.44% (341) | arsenic-transport integral membrane protein ArsC |
| M. avium 104 | MAV_1495 | arsB | 1e-176 | 85.92% (348) | arsenical-resistance protein |
| M. thermoresistible (build 8) | TH_1048 | arsC | 1e-175 | 88.20% (339) | PROBABLE ARSENIC-TRANSPORT INTEGRAL MEMBRANE PROTEIN ARSC |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3222 | - | 1e-178 | 88.32% (351) | arsenical-resistance protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2676|M.bovis_AF2122/97 MTETVTRTAAPAVVGKLSTLDRFLPVWIGSAMAAGLLLGRWIPGLHTALE
Rv2643|M.tuberculosis_H37Rv MTETVTRTAAPAVVGKLSTLDRFLPVWIGSAMAAGLLLGRWIPGLHTALE
MMAR_2055|M.marinum_M MTGTVDHQPAPAVVGKLSTLDRFLPVWIGLAMAAGLLLGRWIPGLNTALE
TH_1048|M.thermoresistible__bu ----------------MSVLDRYLTVWIGAAMIAGLLLGRWIPGLNTALE
Mvan_3222|M.vanbaalenii_PYR-1 --MSDTSTASPAVVAKLSTLDRFLPLWIGAAMVAGLLLGRWIPGLNTALE
MSMEG_1172|M.smegmatis_MC2_155 MTTTSTTAT---VVGKLSTLDRFLPVWIGAAMAAGLLLGRWIPGLNTALE
MAB_4863|M.abscessus_ATCC_1997 MNATADTAEHPAVAGKLSTLDRFLPVWIGVAMAAGLLLGRNVPGLNTALE
MAV_1495|M.avium_104 -MNSPTTTDPTPVVGKLSTLDRFLPVWIGAAMVAGLLLGRAVPGLDRALS
Mflv_3469|M.gilvum_PYR-GCK MSQQVESGPDHAVVGKLSTLDRFLPVWIIAAMIAGLLLGRMVPGFDTALN
:*.***:*.:** ** ******* :**:. **.
Mb2676|M.bovis_AF2122/97 GVQLDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLLSSLLLNWV
Rv2643|M.tuberculosis_H37Rv GVQLDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLLSSLLLNWV
MMAR_2055|M.marinum_M KVQVDGISLPIALGLLIMMYPVLAKVRYDRLDSVTGDRKLLISSLLLNWV
TH_1048|M.thermoresistible__bu SVAIDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRRLLLSSLVLNWV
Mvan_3222|M.vanbaalenii_PYR-1 SVAIDGVSLPIALGLLIMMYPVLAKVRYDRLDTVTGDRRLLISSLVLNWV
MSMEG_1172|M.smegmatis_MC2_155 SIQVDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLASSLILNWA
MAB_4863|M.abscessus_ATCC_1997 KVQVDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLLGSLVLNWV
MAV_1495|M.avium_104 AVQLDGISLPIAIGLLVMMYPVLAKVRYDRLDAVTGDRKLMVASLVLNWV
Mflv_3469|M.gilvum_PYR-GCK SAQVQGISIPIAVGLLVMMYPVLAKVRYDRLGTVTGDRRLLVSSLVLNWV
::*:*:***:***:**************.:*****:*: .**:***.
Mb2676|M.bovis_AF2122/97 LGPALMFALAWLLLADLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
Rv2643|M.tuberculosis_H37Rv LGPALMFALAWLLLADLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
MMAR_2055|M.marinum_M AGPALMFALAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLARGDREAA
TH_1048|M.thermoresistible__bu FGPALMFALAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
Mvan_3222|M.vanbaalenii_PYR-1 FGPALMFALAWLMLPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
MSMEG_1172|M.smegmatis_MC2_155 LGPALMFTLAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
MAB_4863|M.abscessus_ATCC_1997 LGPALMFALAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
MAV_1495|M.avium_104 IGPALMFALAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
Mflv_3469|M.gilvum_PYR-GCK LGPALMFALAWLLVPDLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
******:****::.**************************** ******
Mb2676|M.bovis_AF2122/97 AVLVALNSIFQVAMFAALGWFYLSVLPGWLGLEQTTIATSPWQIAKSVLI
Rv2643|M.tuberculosis_H37Rv AVLVALNSIFQVAMFAALGWFYLSVLPGWLGLEQTTIATSPWQIAKSVLI
MMAR_2055|M.marinum_M AVLVALNSIFQVAMFAVLGWFYLSVLPGWLHLTQTTISTSPWQIAKSVLI
TH_1048|M.thermoresistible__bu AVLVALNSIFQVIMFAVLGWFYLSVLPGWLGLEQTTIDTSPWQIAKSVLI
Mvan_3222|M.vanbaalenii_PYR-1 AVLVALNSIFQVFMFAVLGWFYLSVLPGWLGLEQTTIDTSPWQIAKSVLI
MSMEG_1172|M.smegmatis_MC2_155 AVLVALNSIFQVVMFAVLGWFYLSVLPGWLGLEQATITTSPWQIARSVLI
MAB_4863|M.abscessus_ATCC_1997 AVLVALNSVFQVVMFAVLGWFYLSVLPGWLHLPQTEISTSPWQIAKSVLI
MAV_1495|M.avium_104 AVLVALNSVFQVVMFAVLGWFYLSVLPGWLGLDQTSISVSPWQIAKSVLI
Mflv_3469|M.gilvum_PYR-GCK AVLVALNSLFQVVMFAVLGWFYLAVLPGWLGLETASIDASPWEIAQSVLI
********:*** ***.******:****** * : * .***:**:****
Mb2676|M.bovis_AF2122/97 FLGIPLLAGYLSRRIGEKTKGRNWYESRFLPKVGPWALYGLLFTIVILFA
Rv2643|M.tuberculosis_H37Rv FLGIPLLAGYLSRRIGEKTKGRNWYESRFLPKVGPWALYGLLFTIVILFA
MMAR_2055|M.marinum_M FLGIPLLAGYLSRRLGERAKGRGWYESRFLPKVGPWALYGLLFTIVILFA
TH_1048|M.thermoresistible__bu FLGIPLLAGYLSRRVGERTKGRDWYESIFLPRVSPWALYGLLFTVVILFA
Mvan_3222|M.vanbaalenii_PYR-1 FLGIPLLAGYLSRRIGEKTKGRAWYESRFLPVIGPWALYGLLFTIVILFA
MSMEG_1172|M.smegmatis_MC2_155 FLGIPLAAGYLSRRIGEKARGREWYESRFLPRVGPWALYGLLFTIVVLFA
MAB_4863|M.abscessus_ATCC_1997 FLGIPLLAGYLSRSIGERTRGRDWYESKFLPVIGPWALYGLLFTIVILFA
MAV_1495|M.avium_104 FLGVPLLLGYLSRRLGERTKGRDWYEDRFLPKVGPWALYGLLFTIVILFA
Mflv_3469|M.gilvum_PYR-GCK FLGIPLLAGYLTRRLGEKAKGRTWYETRFLPRIGPWALYGLLFTIVVLFA
***:** ***:* :**:::** *** *** :.**********:*:***
Mb2676|M.bovis_AF2122/97 LQGDQITGRPLDVARIALPLLAYFAIMWVGGYLLGAALRLGYRRTTTLAF
Rv2643|M.tuberculosis_H37Rv LQGDQITGRPLDVARIALPLLAYFAIMWVGGYLLGAALRLGYRRTTTLAF
MMAR_2055|M.marinum_M LQGDQITNRPQDVARIALPLLVYFAVMWGGGYLLGAALHLGYQRTTTMAF
TH_1048|M.thermoresistible__bu LQGDQITNRPLDVTRIALPLLAYFTIMWGGGYLTGAALGLGYERTTTLAF
Mvan_3222|M.vanbaalenii_PYR-1 LQGDQITNRPWDVARIALPLLVYFAVMWGGGYALGAALRLGYERTTTLAF
MSMEG_1172|M.smegmatis_MC2_155 LQGEQITSRPLDVVRIALPLLVYFAIMWGGGYLLGAVLGLSYERTTTLAF
MAB_4863|M.abscessus_ATCC_1997 LQGDQITSKPWDVARIALPLLAYFAIMWGGGYLLGAAMGLGYARTTTLAF
MAV_1495|M.avium_104 LQGAQITSRPLDVVRIAVPLLVYFAVMWGGGYALGAVLGLGYQRTTTLAF
Mflv_3469|M.gilvum_PYR-GCK LQGQQITSRPLDVARIALPLLVYFAVMWGGGYLLGAALGLGYRRTTTLAF
*** ***.:* **.***:***.**::** *** **.: *.* ****:**
Mb2676|M.bovis_AF2122/97 TAASNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
Rv2643|M.tuberculosis_H37Rv TAASNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
MMAR_2055|M.marinum_M TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
TH_1048|M.thermoresistible__bu TAAGNNFELAIAVAIATYGATSGQALATVVGPLIEVPVLVALIYVSLALR
Mvan_3222|M.vanbaalenii_PYR-1 TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVALVYVSLALR
MSMEG_1172|M.smegmatis_MC2_155 TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVALVYVSLALR
MAB_4863|M.abscessus_ATCC_1997 TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVALVYVSLALR
MAV_1495|M.avium_104 TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVALVYVSLALR
Mflv_3469|M.gilvum_PYR-GCK TAAGNNFELAIAVCIATYGATSGQALAGVVGPLIEVPVLVALVYVSLFLR
***.*********.************* ************.*:**** **
Mb2676|M.bovis_AF2122/97 NRLAGPN-ATHDADKPSVLFVCVHNAGRSQMAAGLLTHLAGDRIEVRSAG
Rv2643|M.tuberculosis_H37Rv NRLAGPN-ATHDADKPSVLFVCVHNAGRSQMAAGLLTHLAGDRIEVRSAG
MMAR_2055|M.marinum_M NRYYGPHRRKHDMIKPSVLFVCVHNAGRSQMAAGWLRHLAGDRIEVRSAG
TH_1048|M.thermoresistible__bu QRFERTP-----------------------LAP-----------------
Mvan_3222|M.vanbaalenii_PYR-1 RRFPDST-----------------------TDSTRTKEVVDD--------
MSMEG_1172|M.smegmatis_MC2_155 KRFNRAK-----------------------EHTS----------------
MAB_4863|M.abscessus_ATCC_1997 GRFTPAP-----------------------QHNSPAASTAASTTNVTE--
MAV_1495|M.avium_104 RRFKSST-----------------------PESIPTTDRNTP--------
Mflv_3469|M.gilvum_PYR-GCK RRFHAGS------------------------DNRPREDATDG--------
*
Mb2676|M.bovis_AF2122/97 TEPAGQVNPTAVAAMAEMGIDITANAPTLLTGGQVQSSDVVITMGCGDAC
Rv2643|M.tuberculosis_H37Rv TEPAGQVNPTAVAAMAEMGIDITANAPTLLTGGQVQSSDVVITMGCGDAC
MMAR_2055|M.marinum_M TEPADRINPVAVAAMAELGIDITANTPEVLTGAWVQTSDVVITMGCGDAC
TH_1048|M.thermoresistible__bu --------------------------------------------------
Mvan_3222|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1172|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4863|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1495|M.avium_104 --------------------------------------------------
Mflv_3469|M.gilvum_PYR-GCK --------------------------------------------------
Mb2676|M.bovis_AF2122/97 PYFPGASYRNWKLPDPAGQPLDVVRMIRDDIADRVQALIAELLATAKTR-
Rv2643|M.tuberculosis_H37Rv PYFPGVSYRNWKLPDPAGQPLDVVRMIRDDIADRVQALIAELLATAKTR-
MMAR_2055|M.marinum_M PYFPGVSYRDWTLADPAGQPLDVVRPIRDDIGERVRALIAELLNEHPDTA
TH_1048|M.thermoresistible__bu --------------------------------------------------
Mvan_3222|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1172|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4863|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1495|M.avium_104 --------------------------------------------------
Mflv_3469|M.gilvum_PYR-GCK --------------------------------------------------
Mb2676|M.bovis_AF2122/97 -----
Rv2643|M.tuberculosis_H37Rv -----
MMAR_2055|M.marinum_M QICRG
TH_1048|M.thermoresistible__bu -----
Mvan_3222|M.vanbaalenii_PYR-1 -----
MSMEG_1172|M.smegmatis_MC2_155 -----
MAB_4863|M.abscessus_ATCC_1997 -----
MAV_1495|M.avium_104 -----
Mflv_3469|M.gilvum_PYR-GCK -----