For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDVLERLDLDTVRDLSQRTDRLGVVSLYFNADPTQNLQVQAIDLRNRYRLLQRRMEADGSQRYRDVAAAL ERRRPLIETLITGVASGRGRIMFIGLDGDWTVHFECMMPVTNRVVLDDGPFIHPLVEVLDEGRPAGVVVI SADDARLLEWRLGQLQEISRIAQPFGQAPQERAGQMREQRQAREREGMQRFLTQVTATATELAEERGWER IVVSGGQRWTEPAKTRFGPPLRGKVLVDRGLGGLDDAALTAAVSARLHRQHKYDEQQLLDYLCDAGGTGD VALGLSEVTAALNAGRVDHLVYDPAVRYVGTVGADGSLYADGEIAPHSHAATKDSRFTERLVDRALATGA RISPVEGAADAVLKDAAGVAAVLRW
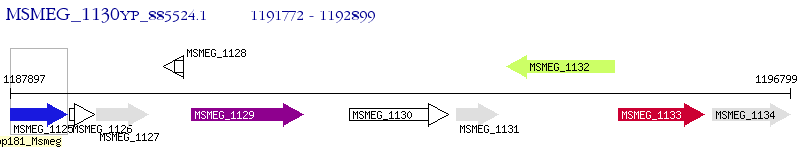
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1130 | - | - | 100% (375) | hypothetical protein MSMEG_1130 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0139 | - | 1e-118 | 57.95% (390) | hypothetical protein MSMEG_1130 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1130|M.smegmatis_MC2_155 MDVLERLDLDTVRDLSQRTDRLGVVSLYFNADP--TQNLQVQAIDLRNRY
TH_0139|M.thermoresistible__bu VVALERLDADSLRDLSRRTDKIGVLSVYVDADPKPDPNLSGVATDLKNRF
: .***** *::****:***::**:*:*.:*** **. * **:**:
MSMEG_1130|M.smegmatis_MC2_155 RLLQRRMEADGSQRYRDVAAALERRRPLIETLITGVASGRGRIMFIGLDG
TH_0139|M.thermoresistible__bu RELQRRMAEDKSERGRDVTAALERVWTQIEWMTSPMFPGRGRVAFFGLGD
* ***** * *:* ***:***** . ** : : : .****: *:**..
MSMEG_1130|M.smegmatis_MC2_155 DWTVHFECMMPVTNRVVLDDGPFIHPLVEVLDEGRPAGVVVISADDARLL
TH_0139|M.thermoresistible__bu GWTERHEIAMPVANRLVLDDGPFIHPLLEALDEGRPAGVIILATQEARLL
.** :.* ***:**:***********:*.*********:::::::****
MSMEG_1130|M.smegmatis_MC2_155 EWRLGQLQEISRIAQPFGQAPQERAGQM------------REQRQARERE
TH_0139|M.thermoresistible__bu EWRVGTLQTVSVMEQPYVEAPHERAGQIGGGPPGQFNAPVREQRQARERV
***:* ** :* : **: :**:*****: *********
MSMEG_1130|M.smegmatis_MC2_155 GMQRFLTQVTATATELAEERGWERIVVSGGQRWTEPAKTRFGPPLRGKVL
TH_0139|M.thermoresistible__bu LTERFVEEIIGVATELAGQRGWERILLSGGPRWTESAVAKFPPPLRDNVF
:**: :: ..***** :******::*** ****.* ::* ****.:*:
MSMEG_1130|M.smegmatis_MC2_155 VD-RGLGGLDDAALTAAVSARLHRQHKYDEQQLLDYLCDAGGTGDVALGL
TH_0139|M.thermoresistible__bu GDTRVLSGLDDAELAATVTEWAHEQHREREKQLVDRIREATGHRRSALGL
* * *.***** *:*:*: *.**: *:**:* : :* * ****
MSMEG_1130|M.smegmatis_MC2_155 SEVTAALNAGRVDHLVYDPAVRYVGTVGADGSLYADGEIAPHSHAATKDS
TH_0139|M.thermoresistible__bu SEVAGALNAGRVAHLVYDPEVRYVGSVGADGSLYAGDEVGPTG-EASKDP
***:.******* ****** *****:*********..*:.* . *:**.
MSMEG_1130|M.smegmatis_MC2_155 RFTERLVDRALATGARISPVEGAADAVLKDAAGVAAVLRW
TH_0139|M.thermoresistible__bu RFTERLIERALETRATVSPVEGAAKGELRDAEGVAALLRW
******::*** * * :*******.. *:** ****:***