For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GKIVTIDYIDDLDGVPIDEKDVDTVEFSYRGEDYTLVLTTKNGAQFNKDIARYIKAAKKAKADDGRVTRK AARARKPAEAKAASKAESNGTSNGVSKRASNRALKAAAAKGAPKRKASSRRTASKTGSSQLRV*
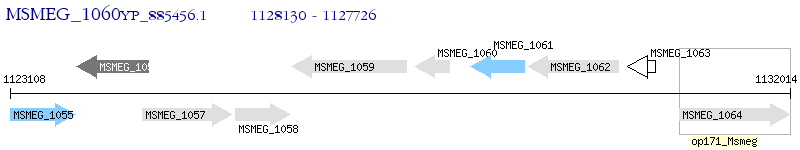
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1060 | - | - | 100% (134) | putative Lsr2 protein |
| M. smegmatis MC2 155 | MSMEG_6092 | - | 1e-05 | 27.45% (102) | Lsr2 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3628c | lsr2 | 5e-05 | 28.00% (100) | LSR2 protein precursor |
| M. gilvum PYR-GCK | Mflv_1429 | - | 2e-05 | 27.62% (105) | Lsr2 |
| M. tuberculosis H37Rv | Rv3597c | lsr2 | 6e-05 | 28.00% (100) | iron-regulated LSR2 protein precursor |
| M. leprae Br4923 | MLBr_00234 | lsr2 | 3e-06 | 29.00% (100) | lsr2 protein (15 kDa antigen) |
| M. abscessus ATCC 19977 | MAB_0545 | - | 5e-05 | 26.80% (97) | protein lsr2 precursor |
| M. marinum M | MMAR_3834 | plsC | 4e-06 | 37.50% (72) | bifunctional transmembrane phospholipid biosynthesis enzyme |
| M. avium 104 | MAV_0555 | - | 3e-05 | 28.12% (96) | Lsr2 protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3764 | plsC | 2e-05 | 38.24% (68) | bifunctional transmembrane phospholipid biosynthesis enzyme |
| M. vanbaalenii PYR-1 | Mvan_5359 | - | 4e-05 | 26.26% (99) | Lsr2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M MSAAEEPGDGREQDKAAGDLRLPGSVAEIMASPAGPKVGAFFDLDGTLVA
MUL_3764|M.ulcerans_Agy99 MSAAEEPGDGREQDKAAGDLRLPGSVAEIMASPAGPKVGAFFDLDGTLVA
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M GFTAVILTQERLRRRDIGVGELLSMVQAGLNHTLGRIEFEQLINTASSAL
MUL_3764|M.ulcerans_Agy99 GFTAVILTQERLRRRDIGVGELLSMVQAGLNHTLGRIEFEQLINTASSAL
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M RGRQLVDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
MUL_3764|M.ulcerans_Agy99 RGRQLVDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 ------------------------------------------------MA
Rv3597c|M.tuberculosis_H37Rv ------------------------------------------------MA
MLBr_00234|M.leprae_Br4923 ------------------------------------------------MA
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK ------------------------------------------------MA
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------MAHWCSRWGVCVHLRRTKGSRLMA
MAB_0545|M.abscessus_ATCC_1997 ------------------------------------------------MA
MMAR_3834|M.marinum_M IQVNPVARFLGIVNMLTNKFETNEDGLLTGGVVKPILWGPGKAAAVQRFA
MUL_3764|M.ulcerans_Agy99 IQVNPVARFLGIVNMLTNKFETNEDGLLTGGVVKPILWGPGKAAAVQRFA
MSMEG_1060|M.smegmatis_MC2_155 ------------------------------------------------MG
Mb3628c|M.bovis_AF2122/97 KKVTVTLVDDFDGSGAAD---ETVEFGLDGVTYEID--------------
Rv3597c|M.tuberculosis_H37Rv KKVTVTLVDDFDGSGAAD---ETVEFGLDGVTYEID--------------
MLBr_00234|M.leprae_Br4923 KKVTVTLVDDFDGAGAAD---ETVEFGLDGVTYEID--------------
MAV_0555|M.avium_104 --MTVTLVDDFDGAGAAD---ETVEFGLDGVTYEID--------------
Mflv_1429|M.gilvum_PYR-GCK KKVTVTLVDDFDGEGGAD---ETVEFGLDGVSYEID--------------
Mvan_5359|M.vanbaalenii_PYR-1 KKVTVTLVDDFDGEGAAD---ETVEFGLDGVSYEID--------------
MAB_0545|M.abscessus_ATCC_1997 KKVTVTLVDDVDGEAPAD---ETVEFGVDGVTYEID--------------
MMAR_3834|M.marinum_M AEHDIDLKDSYFYADGDE---DVALMYLVGNPRPTNPEGKMAAVAKRRGW
MUL_3764|M.ulcerans_Agy99 AEHDIDLKDSYFYADGDE---DVALMYLVGNPRPTNPEGKMAAVAKRRGW
MSMEG_1060|M.smegmatis_MC2_155 KIVTIDYIDDLDGVPIDEKDVDTVEFSYRGEDYTLV--------------
: *. : :.. : *
Mb3628c|M.bovis_AF2122/97 ----LSTKNATKLRGDLKQWV-------------AAGRRVGGRRRGR---
Rv3597c|M.tuberculosis_H37Rv ----LSTKNATKLRGDLKQWV-------------AAGRRVGGRRRGR---
MLBr_00234|M.leprae_Br4923 ----LTNKNAAKLRGDLRQWV-------------SAGRRVGGRRRGR---
MAV_0555|M.avium_104 ----LSSKNAAKLRNDLKQWV-------------EAGRRVGGRRRGR---
Mflv_1429|M.gilvum_PYR-GCK ----LSAKNAAKLRNDLKQWV-------------EAGRRVGGRRRGR---
Mvan_5359|M.vanbaalenii_PYR-1 ----LSAKNAAKLRTDLKQWV-------------EAGRRVGGRRRGR---
MAB_0545|M.abscessus_ATCC_1997 ----LSSKNAEKLRNQLSTWV-------------EHARRVSGRRRGR---
MMAR_3834|M.marinum_M PILKFNSRGGVGLRRQIRTLAGFSTMFPVAAGAVGIGLLTGNRRRGVNFF
MUL_3764|M.ulcerans_Agy99 PILKFNSRGGVGLRRQIRTLAGFSTMFPVAAGAVGIGLLTGNRRRGVNFF
MSMEG_1060|M.smegmatis_MC2_155 ----LTTKNGAQFNKDIARYI--------------KAAKKAKADDGR---
:. :.. :. :: . . *
Mb3628c|M.bovis_AF2122/97 -------SGS--GRGRGAIDREQSAAIR----------------------
Rv3597c|M.tuberculosis_H37Rv -------SGS--GRGRGAIDREQSAAIR----------------------
MLBr_00234|M.leprae_Br4923 -------SNS--GRGRGAIDREQSAAIR----------------------
MAV_0555|M.avium_104 -------SGS--GRGRGAIDREQSAAIR----------------------
Mflv_1429|M.gilvum_PYR-GCK -------AVAGTGRGRAAIDREQSAAIR----------------------
Mvan_5359|M.vanbaalenii_PYR-1 -------AVG-NGRGRAAIDREQSAAIR----------------------
MAB_0545|M.abscessus_ATCC_1997 -------GSSGSGRGRASIDREQSAAIR----------------------
MMAR_3834|M.marinum_M TSNFSQLLLATSGVHLNVVGKENLTAQRPAVFIFNHRNQVDPVIAGALVR
MUL_3764|M.ulcerans_Agy99 TSNFSQLLLATSGVHLNVVGKENLTAQRPAVFIFNHRNQVDPVIAGALVR
MSMEG_1060|M.smegmatis_MC2_155 -------VTRKAARARKPAEAKAASKAES---------------------
. : : .
Mb3628c|M.bovis_AF2122/97 -EWA---RRNGHNVSTRGRI-------------PADVIDAYHAAT-----
Rv3597c|M.tuberculosis_H37Rv -EWA---RRNGHNVSTRGRI-------------PADVIDAYHAAT-----
MLBr_00234|M.leprae_Br4923 -EWA---RRNGHNVSTRGRI-------------PADVIDAFHAAT-----
MAV_0555|M.avium_104 -EWA---RRNGHNVSTRGRI-------------PADVIDAFHAAT-----
Mflv_1429|M.gilvum_PYR-GCK -EWA---RRNGHNVSARGRI-------------PADVIDAFHAAT-----
Mvan_5359|M.vanbaalenii_PYR-1 -DWA---RRNGHNVSTRGRI-------------PADVIDAFHAAT-----
MAB_0545|M.abscessus_ATCC_1997 -EWA---RKNGHNVSSRGRI-------------PAEVVDAFNAAN-----
MMAR_3834|M.marinum_M DNWVGVGKKELQNDPIMGTLGKVLDGVFIDRDDPASAVETLHTVEERAKN
MUL_3764|M.ulcerans_Agy99 DNWVGVGKKELQNDPIMGTLGKVLDGVFIDRDDPASAVETLHTVEERAKN
MSMEG_1060|M.smegmatis_MC2_155 NGTSNGVSKRASNRALKAAAAKG---------APKRKASSRRTASKTGSS
:. * . . * .: .:.
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M GLSIVIAPEGTRLDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAEIVASR
MUL_3764|M.ulcerans_Agy99 GLSIVIAPEGTRLDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAEIVASR
MSMEG_1060|M.smegmatis_MC2_155 QLRV----------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M NSTTINPGTVDIAVFPPIPVDDWTLDTLPDRIAEVRQLYLDTLKDWPVDE
MUL_3764|M.ulcerans_Agy99 NSTTINPGTVDIAVFPPIPVDDWTLDTLPDRIAEVRQLYLDTLKDWPVDE
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M LPEVNLYAEEKAAKKAKAQLAKASAKETPAKKAPAKKAPAKKAAAKKETS
MUL_3764|M.ulcerans_Agy99 LPEVNLYAEEKAAKKAKAQLAKASAKET-----PAKKAPAKKAAAKKET-
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M AKKAPAKKAPAKKAAAKKAPAKKATAKKAPAKKAPAKKVPESAAAKAATP
MUL_3764|M.ulcerans_Agy99 ----PAKKAPAKKAPAKKAPAKKATAKKAPANKAPAKKVPESAAAKAATR
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 --------------------------------------------------
Rv3597c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00234|M.leprae_Br4923 --------------------------------------------------
MAV_0555|M.avium_104 --------------------------------------------------
Mflv_1429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5359|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0545|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3834|M.marinum_M ATDTELPAPKPVGEASNPTEIAPEPIAHDGDAQQLGAGRSDAAESSSSRP
MUL_3764|M.ulcerans_Agy99 ATDTELSAPKPVGEASNPTEIAPEPIAHDGDAQQLGAGRSDAAESSSSRP
MSMEG_1060|M.smegmatis_MC2_155 --------------------------------------------------
Mb3628c|M.bovis_AF2122/97 ----
Rv3597c|M.tuberculosis_H37Rv ----
MLBr_00234|M.leprae_Br4923 ----
MAV_0555|M.avium_104 ----
Mflv_1429|M.gilvum_PYR-GCK ----
Mvan_5359|M.vanbaalenii_PYR-1 ----
MAB_0545|M.abscessus_ATCC_1997 ----
MMAR_3834|M.marinum_M KGRP
MUL_3764|M.ulcerans_Agy99 KGRP
MSMEG_1060|M.smegmatis_MC2_155 ----