For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEFLDLINSYVWSSWLVYLCLAAGVYFSIRTRLLQIRQVPEMIRLMVKGEKSPSGVSSFQALTMSLAGRV GTGNIAGVATAIAFGGPGAMFWMWAVAFLGASTSFVECTLGQIYKARDQLTGEYRGGPAYYLSRALEHTR AAGVFKVYGLLFAAVTVLACGLLLPSVQSNSMAAAMNSAWGFSTWWVGVGIVIVLAFVIIGGVKRIAAFA SIVVPFMAVVYIVLALIIVVVNVSAVPEVLRLIFASAFGLDSAFGAIIGSAVMWGVKRGIYSNEAGQGTG PHAAAAAEVTHPAKQGLVQAFAVYIDTLFVCSATGFLILTTGAYRVYEGESETGAVLAEGGMLPSDITVG PAFAQAGFDTLWSGAGASFVAVSLAFFAFTTIVAYYYMAETNLRFLLGKSATIPVPIVRGTVGSNATMLL QALILVSVVTGAISTATDAWTLGDIGVGLMAWLNIIGIIILQRPALAALKDYEKQQKAGIDPVFNPLSLD IVGATFWETYDPKGRHERTTTSA*
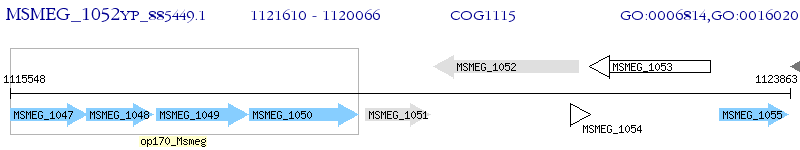
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1052 | - | - | 100% (514) | amino acid carrier protein |
| M. smegmatis MC2 155 | MSMEG_2332 | - | 0.0 | 100.00% (514) | amino acid carrier protein |
| M. smegmatis MC2 155 | MSMEG_0812 | - | 1e-05 | 24.71% (437) | amino acid transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4266 | - | 0.0 | 80.51% (513) | amino acid carrier protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2093 | - | 0.0 | 80.97% (515) | amino acid carrier protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4266|M.gilvum_PYR-GCK MSEFLSTLNDLVWHETLVYLCLAAGVYFSARSRFVQVRQIPEMIRLMMRG
Mvan_2093|M.vanbaalenii_PYR-1 MTEFLADVNDLVWHETLVYLCLGAGVYFSVRSRFVQVRQIPEMVRLMLKG
MSMEG_1052|M.smegmatis_MC2_155 MSEFLDLINSYVWSSWLVYLCLAAGVYFSIRTRLLQIRQVPEMIRLMVKG
*:*** :*. ** . ******.****** *:*::*:**:***:***::*
Mflv_4266|M.gilvum_PYR-GCK EKSPSGVSSFQALTMSLAGRVGTGNIAGVATAIAFGGPGALFWMWMVAFL
Mvan_2093|M.vanbaalenii_PYR-1 EKSPSGVSSFQALTMSLAGRVGTGNIAGVATAIAFGGPGALFWMWAVAFL
MSMEG_1052|M.smegmatis_MC2_155 EKSPSGVSSFQALTMSLAGRVGTGNIAGVATAIAFGGPGAMFWMWAVAFL
****************************************:**** ****
Mflv_4266|M.gilvum_PYR-GCK GASTSFVECTLGQIYKDRDKLTGEYRGGPAYYLSTALSHTAAAGAMKVYG
Mvan_2093|M.vanbaalenii_PYR-1 GASTSFVECTLGQIYKDRDKLTGEYRGGPAYYLSTALSHTAAAGAMKVYG
MSMEG_1052|M.smegmatis_MC2_155 GASTSFVECTLGQIYKARDQLTGEYRGGPAYYLSRALEHTRAAGVFKVYG
**************** **:************** **.** ***.:****
Mflv_4266|M.gilvum_PYR-GCK YVFAAVTLLAMGLLLPSVQSNSMASAMNSAWGFSKWWVAVGTVIVLAFVI
Mvan_2093|M.vanbaalenii_PYR-1 YIFAAVTLLAMGLLLPSVQSNSMASAMNSAWGFSKWWVAVGTVIVLAFVI
MSMEG_1052|M.smegmatis_MC2_155 LLFAAVTVLACGLLLPSVQSNSMAAAMNSAWGFSTWWVGVGIVIVLAFVI
:*****:** *************:*********.***.** ********
Mflv_4266|M.gilvum_PYR-GCK IGGVKRIATFASVVVPFMAVVYIVLALIIVLTNASEIPAIISLIFSSAFG
Mvan_2093|M.vanbaalenii_PYR-1 IGGVKRIATFASIVVPFMAVVYIVLALIIVLMNAAEIPAIISLIFSSAFG
MSMEG_1052|M.smegmatis_MC2_155 IGGVKRIAAFASIVVPFMAVVYIVLALIIVVVNVSAVPEVLRLIFASAFG
********:***:*****************: *.: :* :: ***:****
Mflv_4266|M.gilvum_PYR-GCK LDSAFGAIIGAAVMWGVKRGIYSNEAGQGTGPHAAAAAEVSHPAKQGLVQ
Mvan_2093|M.vanbaalenii_PYR-1 VDSAFGAIIGSAVMWGVKRGIYSNEAGQGTGPHAAAAAEVSHPAKQGLVQ
MSMEG_1052|M.smegmatis_MC2_155 LDSAFGAIIGSAVMWGVKRGIYSNEAGQGTGPHAAAAAEVTHPAKQGLVQ
:*********:*****************************:*********
Mflv_4266|M.gilvum_PYR-GCK AFAVYIDTLFICSATGFLILSTGAYRVFEGESADGAVISEGGDFLASDAE
Mvan_2093|M.vanbaalenii_PYR-1 AFAVYIDTLFICSATGFLILSTGAYRVFEGESADGAVIREGGNILPAGAE
MSMEG_1052|M.smegmatis_MC2_155 AFAVYIDTLFVCSATGFLILTTGAYRVYEGESETGAVLAEGG-MLPSDIT
**********:*********:******:**** ***: *** :*.:.
Mflv_4266|M.gilvum_PYR-GCK VGPAFAQAGFDTLWSGAGSSFIAISLAFFCFTTIVAYYYMAETNLRFMLG
Mvan_2093|M.vanbaalenii_PYR-1 VGPAFAQTGFDTLWSGAGASFIAVSLAFFCMTTIIAYYYMAETNLRFLLG
MSMEG_1052|M.smegmatis_MC2_155 VGPAFAQAGFDTLWSGAGASFVAVSLAFFAFTTIVAYYYMAETNLRFLLG
*******:**********:**:*:*****.:***:************:**
Mflv_4266|M.gilvum_PYR-GCK RFATIDVRFLPGSVGTTMTMLLQALILVSVTLGAVSTASEAWTLGDIGVG
Mvan_2093|M.vanbaalenii_PYR-1 RAATIEVPLIRGTVGSNTTMVLQALILVSVTIGAISTATDAWTLGDIGVG
MSMEG_1052|M.smegmatis_MC2_155 KSATIPVPIVRGTVGSNATMLLQALILVSVVTGAISTATDAWTLGDIGVG
: *** * :: *:**:. **:*********. **:***::**********
Mflv_4266|M.gilvum_PYR-GCK LMAWLNIIGILILQRPAYKALWDYERQKKAGLDPTFDPIALDIRNATFWE
Mvan_2093|M.vanbaalenii_PYR-1 LMAWLNIIGIIILQQPAYKALWDYEKQKKQGLDPMFDPITLDIRNASFWE
MSMEG_1052|M.smegmatis_MC2_155 LMAWLNIIGIIILQRPALAALKDYEKQQKAGIDPVFNPLSLDIVGATFWE
**********:***:** ** ***:*:* *:** *:*::*** .*:***
Mflv_4266|M.gilvum_PYR-GCK NYDPVTGRDKRPTITRA
Mvan_2093|M.vanbaalenii_PYR-1 NYDPATGKDKQPTTTPA
MSMEG_1052|M.smegmatis_MC2_155 TYDPK-GRHERTTTSA-
.*** *:.::.* :