For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAEIKVLVLVGSLRAASINRQLAELAVETAPDGVDLQVFDRLAEVPFYNEDIDTADVPEPAAALRAAAVE ADAALLVTPEYNATIPAVLKNAIDWLSRPYGSGALHGKPAAVIGAAHGRYGGVWAHDETRKSLGVAGPRV VEDVKLSIAASTLDDKHPRENAEVTVSVREAVAKLVAAVAEVPAGVA
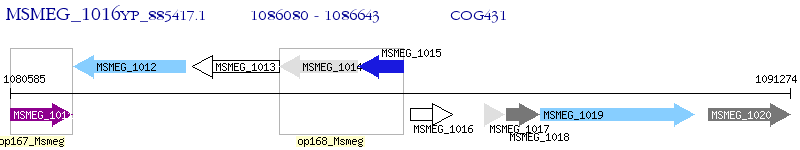
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1016 | - | - | 100% (187) | secreted protein |
| M. smegmatis MC2 155 | MSMEG_2296 | - | e-101 | 100.00% (187) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3080c | - | 1e-58 | 62.57% (179) | hypothetical protein Mb3080c |
| M. gilvum PYR-GCK | Mflv_4308 | - | 2e-69 | 72.41% (174) | NADPH-dependent FMN reductase |
| M. tuberculosis H37Rv | Rv3054c | - | 1e-58 | 62.57% (179) | hypothetical protein Rv3054c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3417c | - | 1e-53 | 56.74% (178) | NADPH-dependent FMN reductase |
| M. marinum M | MMAR_1638 | - | 9e-63 | 63.69% (179) | hypothetical protein MMAR_1638 |
| M. avium 104 | MAV_3922 | - | 2e-58 | 64.57% (175) | secreted protein |
| M. thermoresistible (build 8) | TH_0005 | - | 2e-70 | 74.01% (177) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3398 | - | 3e-62 | 63.13% (179) | hypothetical protein MUL_3398 |
| M. vanbaalenii PYR-1 | Mvan_2039 | - | 1e-70 | 74.14% (174) | NADPH-dependent FMN reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3080c|M.bovis_AF2122/97 ------------MSDTKSDIKILALVGSLRAASFNRQIAELAAKVAPDGV
Rv3054c|M.tuberculosis_H37Rv ------------MSDTKSDIKILALVGSLRAASFNRQIAELAAKVAPDGV
MMAR_1638|M.marinum_M ------------MAQTTSDIKVLALVGSLRVASINRQIAQLAADVAPDGV
MUL_3398|M.ulcerans_Agy99 ------------MAQTTSDIKVLALVGSLRVASINRQIAQLAADVAPDGV
MAV_3922|M.avium_104 -------------------------MGSLRSASINRQIAELASAVAGEDV
Mflv_4308|M.gilvum_PYR-GCK ------------MSENGSST-VLVLVGSLREASVNRQLAELAVESAPADV
Mvan_2039|M.vanbaalenii_PYR-1 MGVSETTIERMKMAQGENNT-VLVLVGSLRAASINRQLAELAVETAPAGV
TH_0005|M.thermoresistible__bu ----VTMADRSEPAAGGADTRVLVLLGSLRAGSINRQLVEVAVETAPAGV
MSMEG_1016|M.smegmatis_MC2_155 ----------------MAEIKVLVLVGSLRAASINRQLAELAVETAPDGV
MAB_3417c|M.abscessus_ATCC_199 ------------MAEKVAEKNVLAIVGSLRKASVNRQLAEAAAEHAPAGV
:**** .*.***:.: * * .*
Mb3080c|M.bovis_AF2122/97 TVTMFEGLGDLPFYNEDID--TATEVP--APVSALREAASDAHAALVVTP
Rv3054c|M.tuberculosis_H37Rv TVTMFEGLGDLPFYNEDID--TATEVP--APVSALREAASDAHAALVVTP
MMAR_1638|M.marinum_M SVTVFEGLGDLPFYNEDID--TPDQVP--APVSALREVAAQSDAALVVTP
MUL_3398|M.ulcerans_Agy99 SVTVFEGLGDLPFYNEDID--TLDQVP--APVSALREVAAQSDAALVVTP
MAV_3922|M.avium_104 VVTVFEGLGELPFYNEEIDDAMTSDAPTLAPVAALRAAAADADAALVVTP
Mflv_4308|M.gilvum_PYR-GCK DVEWFDRLGELPFYNEDID--TAD-VP--EPVAALRTAAARADAALVVTP
Mvan_2039|M.vanbaalenii_PYR-1 ALELFDRLGELPFYNEDLD--TDD-VA--EPVVALREAAARAGAALVITP
TH_0005|M.thermoresistible__bu RLELFDRLGELPFYNEDID--TPGRLA--EPVAALRSAAAEAHAALVVTP
MSMEG_1016|M.smegmatis_MC2_155 DLQVFDRLAEVPFYNEDID--TAD-VP--EPAAALRAAAVEADAALLVTP
MAB_3417c|M.abscessus_ATCC_199 SVTIYEGLADLPHYNEDID---GEGAP--AAAEALRAAVADADAVLLVSP
: :: *.::*.***::* . .. *** .. : *.*:::*
Mb3080c|M.bovis_AF2122/97 EYNGSIPAVIKNAIDWLSRPFGDGALKDKPLAVIGGSMGRYGGVWAHDET
Rv3054c|M.tuberculosis_H37Rv EYNGSIPAVIKNAIDWLSRPFGDGALKDKPLAVIGGSMGRYGGVWAHDET
MMAR_1638|M.marinum_M EYNGSIPAVIKNAIDWLSRPFGDGALKDKPLAVIGGAFGRYGGVWAHDDT
MUL_3398|M.ulcerans_Agy99 EYNGSIPAVIKNAIDWLSRPFGDGALKDKPLAVIGGAFARYGGVWAHDDT
MAV_3922|M.avium_104 EYNGSIPAVIKNAIDWLSRPFGDGALKGKPLAVIGGSFGQYGGVWAHDET
Mflv_4308|M.gilvum_PYR-GCK EYNGTIPGVLKNAIDWLSRPYGDSALKDKPLAVVGAALGQYGGVWAHDET
Mvan_2039|M.vanbaalenii_PYR-1 EYNGTIPGVLKNAIDWLSRPWGNGALKGKPVAVIGAALGQYGGVWAHNET
TH_0005|M.thermoresistible__bu EYNGSIPAVLKNAIDWLSRPYGNGALQGKPAAVIGAALGRYGGKWAHDET
MSMEG_1016|M.smegmatis_MC2_155 EYNATIPAVLKNAIDWLSRPYGSGALHGKPAAVIGAAHGRYGGVWAHDET
MAB_3417c|M.abscessus_ATCC_199 ENNGTISSSLKNAIDWLSRPFGSSVFKGKPTVVIGTSAGRYGGIWAIDET
* *.:*.. :**********:*...::.** .*:* : .:*** ** ::*
Mb3080c|M.bovis_AF2122/97 RKSFSIAGTRVVDAIKLSVPFQTL-GKSVADDAGLAANVRDAVGNLAAEV
Rv3054c|M.tuberculosis_H37Rv RKSFSIAGTRVVDAIKLSVPFQTL-GKSVADDAGLAANVRDAVGNLAAEV
MMAR_1638|M.marinum_M RKSFGIAGARVSDAIKLSVPFLTLEGNPPADNSELTANVRDIIAKLAAEV
MUL_3398|M.ulcerans_Agy99 RKSFGIAGARVSDAIKLSVPFLTLEGNPPADNSELTANVRDIIAKLAAKV
MAV_3922|M.avium_104 RKSFGIAGARVVESIKLSVPFKTLAGKAPAEHAELSANVRDVVGKLAAEV
Mflv_4308|M.gilvum_PYR-GCK RKSFGIAGPRVVDGLELSIPTKTLDGKHPRESDDVVARLRDILGKLVAEI
Mvan_2039|M.vanbaalenii_PYR-1 RKSFGIAGPRVVEDLELSIPTTTLDGKHPREQAEVVARLRDIVGKLAAEI
TH_0005|M.thermoresistible__bu RKSFGIAGPRVLEDPALSLPTKTLDGRHPRENAEVTAAVRTVVEKLAAEA
MSMEG_1016|M.smegmatis_MC2_155 RKSLGVAGPRVVEDVKLSIAASTLDDKHPRENAEVTVSVREAVAKLVAAV
MAB_3417c|M.abscessus_ATCC_199 RKAAGIATANVLEDVKLAVPSSAFDGKHPREHAETVAELTKALDALTTA-
**: .:* ..* : *::. :: .. : . : : *.:
Mb3080c|M.bovis_AF2122/97 G-------
Rv3054c|M.tuberculosis_H37Rv G-------
MMAR_1638|M.marinum_M G-------
MUL_3398|M.ulcerans_Agy99 G-------
MAV_3922|M.avium_104 G-------
Mflv_4308|M.gilvum_PYR-GCK D-------
Mvan_2039|M.vanbaalenii_PYR-1 G-------
TH_0005|M.thermoresistible__bu RVGA----
MSMEG_1016|M.smegmatis_MC2_155 AEVPAGVA
MAB_3417c|M.abscessus_ATCC_199 --------