For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LQTVAIRGYRSLREVVLPLTELTVITGANGTGKSSVYRALRLLADCGRGQVIGSLAREGGLQSVLWAGPE RPSEEAQGATRTRPVSLEMGFAADDFGYVVDLGLPQMAGAPAHRTPSAFTLDPEIKREAVFAGPVLRPSS TLVRRIREFAETAAESGRGFDELSRSLPAYRSVLAEYAHPHALPELSAVSERLRDWRFYDGFRVDAGAPA RHPHVGTRTPVLSDDGSDLAAAVQTIIEAGLDDLQRAVADAFDGARVSVAASDGLFDLQLHQRGMLRPLR AAELSDGTLRFLLWAAALLSPSPPSLMVLNEPETSLHPDLVRPLASLIRTAATRTQVVVVTHSRALLEFL DTTPIGDDGPGGAVEIELYKQWGETKVTGQGLLSTPSWHWGNR*
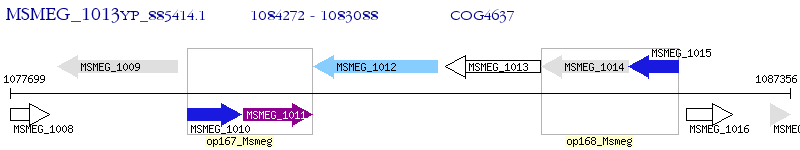
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1013 | - | - | 100% (394) | hypothetical protein MSMEG_1013 |
| M. smegmatis MC2 155 | MSMEG_2293 | - | 0.0 | 100.00% (394) | hypothetical protein MSMEG_2293 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4312 | - | 1e-154 | 70.93% (399) | hypothetical protein Mflv_4312 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01793 | - | 3e-18 | 52.83% (106) | hypothetical protein MLBr_01793 |
| M. abscessus ATCC 19977 | MAB_1548c | - | 1e-145 | 64.71% (408) | hypothetical protein MAB_1548c |
| M. marinum M | MMAR_0146 | - | 1e-105 | 51.00% (402) | ATPase |
| M. avium 104 | MAV_2144 | - | 1e-179 | 79.25% (400) | hypothetical protein MAV_2144 |
| M. thermoresistible (build 8) | TH_0007 | - | 1e-161 | 74.69% (399) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4951 | - | 1e-104 | 50.75% (402) | ATPase |
| M. vanbaalenii PYR-1 | Mvan_2035 | - | 1e-158 | 71.68% (399) | hypothetical protein Mvan_2035 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4312|M.gilvum_PYR-GCK ------------------MLETVAIRGYRSLRDLVLPLRRLTVITGANGT
Mvan_2035|M.vanbaalenii_PYR-1 ------------------MLDTVAIRGYRSLREIVLPLRRLTVITGANGT
MSMEG_1013|M.smegmatis_MC2_155 ------------------MLQTVAIRGYRSLREVVLPLTELTVITGANGT
MAV_2144|M.avium_104 ------MADRTVG----LMLHTVAVRGYRSLREVVLPLGRLSVITGANGT
TH_0007|M.thermoresistible__bu VRRPGRQVDDVIGNDRVSMLQTVAIRGYRSLREVVLPLSRLTVITGANGT
MLBr_01793|M.leprae_Br4923 --------------------------------------------------
MAB_1548c|M.abscessus_ATCC_199 ------------------MLTTVAIRGYRSLRDIVLPLAQLTVITGANGT
MMAR_0146|M.marinum_M ------------------MLVTVAVENYRSLRQLVVPLHQLNVVTGANGS
MUL_4951|M.ulcerans_Agy99 ------------------MLVTVAVENYRSLRQLVVPLHQLNVVTGANGS
Mflv_4312|M.gilvum_PYR-GCK GKSSLYRALRLLADCGRGEVIGSFAREGGVASALWAGPEHLGGARRT--G
Mvan_2035|M.vanbaalenii_PYR-1 GKSSLYRALRLLADCGRGEVIASFAREGGVESAMWAGPEHLGGARRT--G
MSMEG_1013|M.smegmatis_MC2_155 GKSSVYRALRLLADCGRGQVIGSLAREGGLQSVLWAGPERPS-------E
MAV_2144|M.avium_104 GKSSLYRALRLLADCGRGEVIGSLAREGGLQSVLWAGPEQPAGARRS--G
TH_0007|M.thermoresistible__bu GKSSLYRALRLLADCGRGEVIGSLARAGGLQSVLWAGPEHPNADERT--G
MLBr_01793|M.leprae_Br4923 --------------------------------------------------
MAB_1548c|M.abscessus_ATCC_199 GKSSLYRALQLLADCGSGEIIGSLARQGGLQSAMWAGPESLNRARRT--G
MMAR_0146|M.marinum_M GKSNLYRALRLMADSARNGAVAALAREGGLSSTMWAGPAVIGKSVRQGRH
MUL_4951|M.ulcerans_Agy99 GKSNLYRALRLMADSARNGAVAALAREGGLSSTMWAGPAVIGKSVRQGRH
Mflv_4312|M.gilvum_PYR-GCK TAQGGPRSRSVSIELGYAGDDFGYLIDLGLPQATET-------AFGRDPE
Mvan_2035|M.vanbaalenii_PYR-1 TAQGGPRTRPGSIELGYAADDFGYLIDLGLPQARET-------AFGRDPE
MSMEG_1013|M.smegmatis_MC2_155 EAQGATRTRPVSLEMGFAADDFGYVVDLGLPQMAGAPAHRTPSAFTLDPE
MAV_2144|M.avium_104 KTEGTVRTRPVSLELGFAADDFGYLVDLGLPQSAGK------SVFARDPE
TH_0007|M.thermoresistible__bu AAHGTRRTRPVSLELGFAATDFGYLIDLGLPQSAGPG-----SVFARDPE
MLBr_01793|M.leprae_Br4923 --------------------------------------------------
MAB_1548c|M.abscessus_ATCC_199 VVQGTRRTRPVSIELGYASTDFGYLVDLGLPTPSES-------AFGRDPE
MMAR_0146|M.marinum_M QVQGTVRTDSVSLKLGFAGSDFGYAMDLGLPQASIS-------KFKLDPE
MUL_4951|M.ulcerans_Agy99 QVQGTVRTDSVSLKLGFAGSDFSYAMDLGLPQASIS-------KFKLDPE
Mflv_4312|M.gilvum_PYR-GCK IKRELVFAGPVARSTTTLVRRVRGMVEVASDTGRGFDELARNLPAHRSVL
Mvan_2035|M.vanbaalenii_PYR-1 VKRELVFAGPVARPTATLVRRVRGLVEVAADTGRGFDALASNLPAHRSVL
MSMEG_1013|M.smegmatis_MC2_155 IKREAVFAGPVLRPSSTLVRRIREFAETAAESGRGFDELSRSLPAYRSVL
MAV_2144|M.avium_104 IKREVLFAGPVLRPSTTLVRRSRGFAEASADSGSGFDELSRSLPSYHSVL
TH_0007|M.thermoresistible__bu IKREVVFAGPVPRPAATLVRRTRDHAEMRADTG--FEELSRSLPSYHSVL
MLBr_01793|M.leprae_Br4923 --------------------------------------------------
MAB_1548c|M.abscessus_ATCC_199 IKGEAVFTGPSLRPSATLVRRTRPVVEARSESGRGFEGFSRSLPTYRSVL
MMAR_0146|M.marinum_M VKVEALWVGPVWRSATAIAERGGPVARLR-DGDDNWHLITDALRPFDSML
MUL_4951|M.ulcerans_Agy99 VKVEALWVGPVWRSATAIAERGGPVARLR-DGDDNWHLITDALRPFDSML
Mflv_4312|M.gilvum_PYR-GCK TDWAG--ASPELAMVRERLRNWRFYDGFRADADAPARRPQVGTRTPVLAD
Mvan_2035|M.vanbaalenii_PYR-1 ADWAG--AVPELVMVRERLRDWRFYDGFRADSHAPARMPQVGTRTPVLAD
MSMEG_1013|M.smegmatis_MC2_155 AEYAHPHALPELSAVSERLRDWRFYDGFRVDAGAPARHPHVGTRTPVLSD
MAV_2144|M.avium_104 AEYAHPHALPELAAVRDRLRGWRFYDGFRVDAGAPARHPQVGTRTPVLAD
TH_0007|M.thermoresistible__bu AEYAHPEALPELAAVRNRLRRWRFYDGFRVDAAAPARRPQVGTRTPVLSD
MLBr_01793|M.leprae_Br4923 -----------------------------MNATAPARPPQGDTRTPVLAD
MAB_1548c|M.abscessus_ATCC_199 SEFAG--KFPELGAVRDTLRGWRFYDGFRVDALAPARQPQIGTRTTVLSN
MMAR_0146|M.marinum_M SELADPVRAPELIEMRERIRNWRFYDHLRTDADAPARQARIGTRTTVLGH
MUL_4951|M.ulcerans_Agy99 SELADPVRAPELIEMRERIRNWRFYDHLRTDADAPARQARIGTRTTVLGH
:: **** .: .***.**..
Mflv_4312|M.gilvum_PYR-GCK DGSDLAAAIQTILETGNGD-LARAVDAAFDGATVSVAITDGLFDLQLHQR
Mvan_2035|M.vanbaalenii_PYR-1 DGADLAAAVQTILETGTDD-FGRAVADAFDGATVSVSVTDGLFDLHLRQR
MSMEG_1013|M.smegmatis_MC2_155 DGSDLAAAVQTIIEAGLDD-LQRAVADAFDGARVSVAASDGLFDLQLHQR
MAV_2144|M.avium_104 DGSDLAAAIQTILESGFDD-LDRAVAEAFDGATVSVAIHDGLFDLQLRQR
TH_0007|M.thermoresistible__bu DGHDLAAAIATITETGFDD-LARAVGDAFDGATVSVAVHDGLFELQVRQR
MLBr_01793|M.leprae_Br4923 DNSDLAAAIRRSSSQDSMS-GDSPSRTHSMRATVSVVIHDGLFDLQLQQH
MAB_1548c|M.abscessus_ATCC_199 DGSDVAAAIQTIVETGNDT-LDKVVAAAFDGATVAVAVTDGMFNLHVQQP
MMAR_0146|M.marinum_M DGADLAAALETIREIGDEAALDEAIDRAFEGSRIDVASDNGRFELALKQP
MUL_4951|M.ulcerans_Agy99 DGADLAAALETIREIGDEAALDEAIDRAFEGSRIDVASDNGRFELALKQP
*. *:***: . . : : * :* *:* ::*
Mflv_4312|M.gilvum_PYR-GCK GMLRPLRSAEISDGTLRFLLWAAALLSPQPPSLMVLNEPETSLHPDLVPP
Mvan_2035|M.vanbaalenii_PYR-1 GMLRPLRSAEMSDGTLRFLLWAAALLSPQPPSLMVLNEPETSLHPDLVRP
MSMEG_1013|M.smegmatis_MC2_155 GMLRPLRAAELSDGTLRFLLWAAALLSPSPPSLMVLNEPETSLHPDLVRP
MAV_2144|M.avium_104 GMLRALRSAELSDGTLRFLLWAAALLSPQPPSLMVLNEPETSLHPDLVAP
TH_0007|M.thermoresistible__bu GMLRPLRAAELSDGTLRFLLWTAALLSPEPPSLMVLNEPETSLHPELVRP
MLBr_01793|M.leprae_Br4923 EMLR---AAELYDGTLRFLLWTAALFSALALSLI----------------
MAB_1548c|M.abscessus_ATCC_199 GMLRPLRAAELSDGTLRFLLWAAALLSPDPPPLMVLNEPETSLHPDLIAP
MMAR_0146|M.marinum_M GMLRPLGAAELSDGTLRYLLWSAALLSPRPPQLLVLNEPETSLHPELLPA
MUL_4951|M.ulcerans_Agy99 GMLRPLGAAELSDGTLRYLLWSAALLSPRPPQLLVLNEPETSLHPELLPA
*** :**: *****:***:***:*. . *:
Mflv_4312|M.gilvum_PYR-GCK LARLIGEAATRTQLVVVTHSAMLRECLGAEPIGTDGD-----------AW
Mvan_2035|M.vanbaalenii_PYR-1 LAELIDAAATRTQVVVVTHSATLRTNLGAQPIGSDGD-----------AW
MSMEG_1013|M.smegmatis_MC2_155 LASLIRTAATRTQVVVVTHSRALLEFLDTTPIGDDGP---------GGAV
MAV_2144|M.avium_104 LASLIRTTAARTQVVVVTHSRSLLEFLDTVPLGEGGAD--------GEAV
TH_0007|M.thermoresistible__bu VANLIRAAAQRTQLVVVTHSAALLELLGDAAEG-----------------
MLBr_01793|M.leprae_Br4923 --------------------------------------------------
MAB_1548c|M.abscessus_ATCC_199 LATLIQAAAAKTQVVVITHASALMSHLAARPLREILAGEEDYFDDAGPAV
MMAR_0146|M.marinum_M LASLIATAAQRSQIIVVTHSEPLVAGLQQAAEVHT---------------
MUL_4951|M.ulcerans_Agy99 LASLIATAAQRSQIIVVTHSEPLVAGLQQAAEVHT---------------
Mflv_4312|M.gilvum_PYR-GCK EIELYKDWGETRVGDQDLSTMPPWDWGRR
Mvan_2035|M.vanbaalenii_PYR-1 EIELYKDWGETKIAEQGLLTTPPWDWGKR
MSMEG_1013|M.smegmatis_MC2_155 EIELYKQWGETKVTGQGLLSTPSWHWGNR
MAV_2144|M.avium_104 EIELYKDWGETRIGGQTLMTTPRWDWGTR
TH_0007|M.thermoresistible__bu -VELYKDRGETKVAGQTMLSTPPWDWGSR
MLBr_01793|M.leprae_Br4923 -----------------------------
MAB_1548c|M.abscessus_ATCC_199 RLELTKDWGETQIIGLDPLKAPSWHWGSR
MMAR_0146|M.marinum_M -IRLTKELGETRIVGQGMLDQPSWHWPPR
MUL_4951|M.ulcerans_Agy99 -IRLTKELGETRIVGQGMLDQPSWHWPPR