For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTPFEQIAPAFVEMAHSIVWASAATVDADGRPRSRVLHPIWEFDGPDLFGWIATVPSPVKRAHLAEHPYM SLNYWAPNHDTCSAECLVEWYTDDETCRLVWDKFANGPAPVGYDPNIIPQWRGGPTSPEFAALRLAPYRL RVMPGTVMTAGEGEPLLWQS
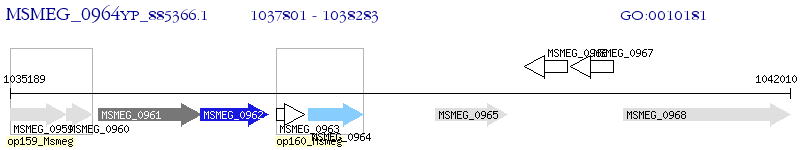
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0964 | - | - | 100% (160) | pyridoxamine 5'-phosphate oxidase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0057 | - | 1e-74 | 77.22% (158) | hypothetical protein Mflv_0057 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2254 | - | 5e-62 | 63.75% (160) | pyridoxamine 5'-phosphate oxidase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0857 | - | 7e-70 | 71.25% (160) | hypothetical protein Mvan_0857 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0057|M.gilvum_PYR-GCK MPSLRAVTALESIAPAFVDMAHSIVWASIATVDAVGRPRTRILHPIWEWD
Mvan_0857|M.vanbaalenii_PYR-1 MPSLASVTPLNRIAPSFVTMAHSIVWASVATVDADSKPRTRILHPYWEWD
MSMEG_0964|M.smegmatis_MC2_155 ------MTPFEQIAPAFVEMAHSIVWASAATVDADGRPRSRVLHPIWEFD
TH_2254|M.thermoresistible__bu ------VTALQRVAPAFVAMAHEIVWATAATVGADGVPRTRILHPLWQWD
:*.:: :**:** ***.****: ***.* . **:*:*** *::*
Mflv_0057|M.gilvum_PYR-GCK GTDLFGWIATVPTPLKRAHLAAHPDVSVSYWTTTQDTCSAECLVEWYTDD
Mvan_0857|M.vanbaalenii_PYR-1 GTDLFGWVATVPSPVKRAHLAVHPDVSVSYWTTTHDTCSAECLVEWYSDD
MSMEG_0964|M.smegmatis_MC2_155 GPDLFGWIATVPSPVKRAHLAEHPYMSLNYWAPNHDTCSAECLVEWYTDD
TH_2254|M.thermoresistible__bu GERLTGWIATVPTPLKRGHIAAHPVVSLSYWSPSHDTCSAECRVRWVYDD
* * **:****:*:**.*:* ** :*:.**:..:******* *.* **
Mflv_0057|M.gilvum_PYR-GCK DTCAQVWDKFATGPAPVGYDPRIIPMWKDGPTSHEFAVLRLAPYRLRVMP
Mvan_0857|M.vanbaalenii_PYR-1 DTRAAIWQKFATAPPPVGYDPSIIPFWKDGPTSDQFAVLRLAPYRLRVMP
MSMEG_0964|M.smegmatis_MC2_155 ETCRLVWDKFANGPAPVGYDPNIIPQWRGGPTSPEFAALRLAPYRLRVMP
TH_2254|M.thermoresistible__bu AGRSTVWELFKNAPGPVGYDPAIIPQWRDGPTCEAFAAWRLTPLRLRVMP
:*: * ..* ****** *** *:.***. **. **:* ******
Mflv_0057|M.gilvum_PYR-GCK GAVMMRGEGAPLMWSDHSGRRD
Mvan_0857|M.vanbaalenii_PYR-1 GTVMTEGAGEIQNWHAS-----
MSMEG_0964|M.smegmatis_MC2_155 GTVMTAGEGEPLLWQS------
TH_2254|M.thermoresistible__bu GTVLTQGRGDVLHWRA------
*:*: * * *