For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MCPSQSPPSEPARAKSLVETVGFVFWLVAAVLLMVGGMIAASVSLPGLDTGLFRGVGVLIAVAGAGMAFL AGRSRNGDPRFRRAAMALSVAIVVIVALTARLGVVHILTLIGIVLLIVGTILNALPRRDRD
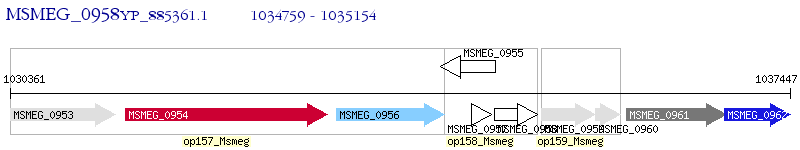
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0958 | - | - | 100% (131) | hypothetical protein MSMEG_0958 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0063 | - | 1e-14 | 30.71% (140) | hypothetical protein Mflv_0063 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3988c | - | 2e-12 | 37.59% (133) | hypothetical protein MAB_3988c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3776 | - | 7e-12 | 33.09% (136) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0849 | - | 2e-12 | 34.92% (126) | hypothetical protein Mvan_0849 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0063|M.gilvum_PYR-GCK MIGYADPTMSAPTSSGRPRIVDLGFWCFMAGAVIMIVGGLMAVTATFEAA
Mvan_0849|M.vanbaalenii_PYR-1 --------MTAPTPSSRPRIVDIAFWCFIAGAVIMIVGGLMALTATYDAA
TH_3776|M.thermoresistible__bu --------VSATSPPQRPRIVDVAFWLLLLGAALLLGTGLVVATVGFDQL
MAB_3988c|M.abscessus_ATCC_199 --------MTAEQP--RPRSVDVAFWFWVVSAAALFLNGLAGVTQRYDAV
MSMEG_0958|M.smegmatis_MC2_155 --MCPSQSPPSEPARAKSLVETVGFVFWLVAAVLLMVGGMIAASVSLPGL
.: . :. :.* : .*. :: *: :
Mflv_0063|M.gilvum_PYR-GCK RAAIPDTVSDDQVRSYLTIYRGTGFGAVLGAGALAFLAGRARRGDARFRL
Mvan_0849|M.vanbaalenii_PYR-1 RSAIPPTVSDEQVRSYLTIYRFSGIGSVLAAGALAFLAGRARRGDARFRL
TH_3776|M.thermoresistible__bu RAVAPETFTDDQLRSYLMFRRATGLLYALAGAGLAFLTGRIRGGQVTYRR
MAB_3988c|M.abscessus_ATCC_199 RAAARHGLTDEDVRNLVTYFRAWGALCILLAAGIAFLAGRTRQGDKRYRR
MSMEG_0958|M.smegmatis_MC2_155 DTG---------------LFRGVGVLIAVAGAGMAFLAGRSRNGDPRFRR
: * * : ...:***:** * *: :*
Mflv_0063|M.gilvum_PYR-GCK ATLALTFAAVVVIGLLALGIGVAQPLILLGLLPAVIGALLFVQPSAREWF
Mvan_0849|M.vanbaalenii_PYR-1 ATLALGFAAVVVVGMLAIGVGVAQPLILLGLLPVLIGALLLVQPSARLWF
TH_3776|M.thermoresistible__bu ATVALGLVTVAVVAVLTVIVGTG-VAPLIALLPVLVGVVLLTRPTAAGWF
MAB_3988c|M.abscessus_ATCC_199 ALIALSVVSVLGAIVMASSGSVG-PLLLIAALSLIVANVLIIRPAAQEWF
MSMEG_0958|M.smegmatis_MC2_155 AAMALSVAIVVIVALTAR-LGVVHILTLIGIVLLIVGTILNALPRRDRD-
* :** .. * : : .. *:. : ::. :* *
Mflv_0063|M.gilvum_PYR-GCK QRDGGS--------
Mvan_0849|M.vanbaalenii_PYR-1 QGFPGKDLS-----
TH_3776|M.thermoresistible__bu TDDVTEDTQDRGEQ
MAB_3988c|M.abscessus_ATCC_199 DGGDHG--------
MSMEG_0958|M.smegmatis_MC2_155 --------------