For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTLLTRAGCHLCARAAEELTVLRDELGFTLVTTDVDALAAEGDNSLRAQYGDLLPVVLLDGTEHSYWEVD AAQLRSDLGH
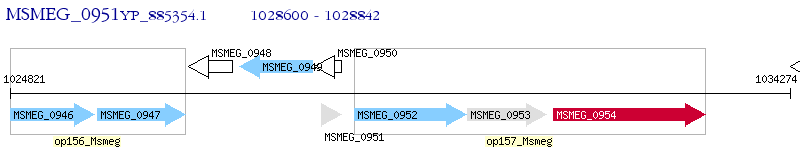
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0951 | - | - | 100% (80) | glutaredoxin 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0520 | - | 6e-24 | 61.54% (78) | hypothetical protein Mb0520 |
| M. gilvum PYR-GCK | Mflv_0068 | - | 8e-27 | 70.51% (78) | glutaredoxin 2 |
| M. tuberculosis H37Rv | Rv0508 | - | 6e-24 | 61.54% (78) | hypothetical protein Rv0508 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3994c | - | 1e-23 | 66.67% (78) | hypothetical protein MAB_3994c |
| M. marinum M | MMAR_0841 | - | 1e-21 | 58.97% (78) | hypothetical protein MMAR_0841 |
| M. avium 104 | MAV_4641 | - | 2e-21 | 58.97% (78) | hypothetical protein MAV_4641 |
| M. thermoresistible (build 8) | TH_0270 | - | 3e-24 | 64.10% (78) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4597 | - | 8e-22 | 58.97% (78) | hypothetical protein MUL_4597 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0068|M.gilvum_PYR-GCK -------MDRQVELLTREGCPMCAAAAAQLDALAGELGFTVVVTDVDAAA
TH_0270|M.thermoresistible__bu -------VESRVVLLSRQGCGMCERAAQQLEELAAELGFEFTVTDVDAAA
Mb0520|M.bovis_AF2122/97 --MS----RPQVELLTRAGCAICVRVAEQLAELSSELGFDMMTIDVDVAA
Rv0508|M.tuberculosis_H37Rv --MS----RPQVELLTRAGCAICVRVAEQLAELSSELGFDMMTIDVDVAA
MMAR_0841|M.marinum_M MPMTQPTHRRQVELLTRDGCAICLRVHAQLAELAGELDFDLVSTDVDVAA
MUL_4597|M.ulcerans_Agy99 --MTQPTHRRQVELLTRDGCAICLRVHAQLAELAGELDFDLVSTDVDVAA
MAV_4641|M.avium_104 --MTSATPRPQVELLTRDGCTICERIHARLVELAAELGFELSTTDVDAAA
MSMEG_0951|M.smegmatis_MC2_155 -----------MTLLTRAGCHLCARAAEELTVLRDELGFTLVTTDVDALA
MAB_3994c|M.abscessus_ATCC_199 --------MTSLTLLTRAGCAACDRAESELAALADEFGVTLTVTDVDEAA
: **:* ** * .* * *:.. . *** *
Mflv_0068|M.gilvum_PYR-GCK AAGDAALRAEYGDRLPVVLLDGVEHSYWEVDEARLRADLSG---------
TH_0270|M.thermoresistible__bu AAGDPALRAEFGDRLPVVLLDGDEHSYWEVDEPRLRADLAGR--------
Mb0520|M.bovis_AF2122/97 STGNPGLRAEFGDRLPVVLLDGREHSYWEVDEHRLRADIARSTFGSPPDK
Rv0508|M.tuberculosis_H37Rv STGNPGLRAEFGDRLPVVLLDGREHSYWEVDEHRLRADIARSTFGSPPDK
MMAR_0841|M.marinum_M SAGNAQLRAEFGDRLPVILLDGREHSYWEVDEPRLRADLAS---------
MUL_4597|M.ulcerans_Agy99 SAGNAQLRAEFGDRLPVILLDGREHSYWEVDEPRLRADLAS---------
MAV_4641|M.avium_104 AADNPGLRAEFGDRLPVILLDGREHSYWDIDEPRLRADLAR---------
MSMEG_0951|M.smegmatis_MC2_155 AEGDNSLRAQYGDLLPVVLLDGTEHSYWEVDAAQLRSDLGH---------
MAB_3994c|M.abscessus_ATCC_199 AT-DSSLRAEFGDRLPVVLLDGQEHSYWEVDEPRLRADLKRDR-------
: : ***::** ***:**** *****::* :**:*:
Mflv_0068|M.gilvum_PYR-GCK ---
TH_0270|M.thermoresistible__bu ---
Mb0520|M.bovis_AF2122/97 RLP
Rv0508|M.tuberculosis_H37Rv RLP
MMAR_0841|M.marinum_M ---
MUL_4597|M.ulcerans_Agy99 ---
MAV_4641|M.avium_104 ---
MSMEG_0951|M.smegmatis_MC2_155 ---
MAB_3994c|M.abscessus_ATCC_199 ---