For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRNEIVSGIAGLVAGHILWLAAITLATDSSRVTRWVLVVAALSFVGGAAAAWVGWRRYQQRSRVWAAFLW CLPVSPVLFSLTLLGVTYL
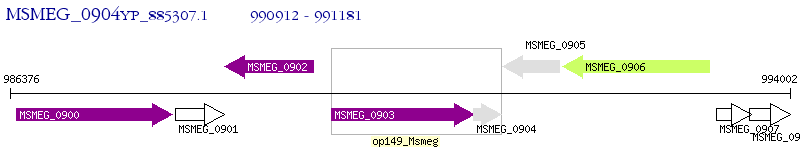
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0904 | - | - | 100% (89) | hypothetical protein MSMEG_0904 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0472 | - | 2e-11 | 36.47% (85) | hypothetical protein Mb0472 |
| M. gilvum PYR-GCK | Mflv_0113 | - | 5e-20 | 56.25% (80) | hypothetical protein Mflv_0113 |
| M. tuberculosis H37Rv | Rv0463 | - | 2e-11 | 36.47% (85) | hypothetical protein Rv0463 |
| M. leprae Br4923 | MLBr_02388 | - | 3e-13 | 35.71% (84) | hypothetical protein MLBr_02388 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0786 | - | 2e-11 | 36.90% (84) | hypothetical protein MMAR_0786 |
| M. avium 104 | MAV_4686 | - | 8e-13 | 44.19% (86) | hypothetical protein MAV_4686 |
| M. thermoresistible (build 8) | TH_4134 | - | 2e-29 | 64.04% (89) | PUTATIVE probable conserved membrane protein |
| M. ulcerans Agy99 | MUL_2215 | - | 1e-11 | 36.90% (84) | hypothetical protein MUL_2215 |
| M. vanbaalenii PYR-1 | Mvan_0795 | - | 4e-22 | 56.52% (92) | hypothetical protein Mvan_0795 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0786|M.marinum_M ----MIR------TEAVLGGAGGVTAGYLLWLVAISIGDDLTTVGQWSLV
MUL_2215|M.ulcerans_Agy99 ----MIR------TEAVLGGAGGVTAGYLLWLVAISIGDDLTTVGQWSLV
Mb0472|M.bovis_AF2122/97 ----MTRRASTDTPQIIMGAIGGVVTGYILWLAAISVGDGLTTVSQWSRV
Rv0463|M.tuberculosis_H37Rv ----MTRRASTDTPQIIMGAIGGVVTGYILWLAAISVGDGLTTVSQWSRV
MLBr_02388|M.leprae_Br4923 MAAGDNRLIGNDTPEIAVGAIGGVATGYVLWLVVMSIGYNITTVSQWSLI
MAV_4686|M.avium_104 ----MLR---TLTPQTIAAALGGLATGYVLWLLAISNGDN-ATVGQWGPL
MSMEG_0904|M.smegmatis_MC2_155 -----MR-------NEIVSGIAGLVAGHILWLAAITLATDSSRVTRWVLV
TH_4134|M.thermoresistible__bu -----LR-------NTVLAGIGGLLLGHTLWLVAISLAMGSSRVNTWVLV
Mflv_0113|M.gilvum_PYR-GCK ------------------------MVGHVLWLAGISFAISSTDISTWVLV
Mvan_0795|M.vanbaalenii_PYR-1 -----MR-------AIWVAGIGGLVVGHILWLAGISAAIASANVPTWVLV
*: *** :: . : : * :
MMAR_0786|M.marinum_M ILLLSGLLAVGAALVGLLMRWRN---RYGWSAFVFGLPVLPVVLTLAVLA
MUL_2215|M.ulcerans_Agy99 ILLLSGLLAVGAALVGLLMRWRN---RYGWSAFVFGLPVLPVVLTLAVLA
Mb0472|M.bovis_AF2122/97 VLLLSVLVAVCGAAGGLRLRSRG---KLAWSAFAFSLPIPPVVLTVAVLA
Rv0463|M.tuberculosis_H37Rv VLLLSVLVAVCGAAGGLRLRSRG---KLAWSAFAFSLPIPPVVLTVAVLA
MLBr_02388|M.leprae_Br4923 VLILSMVSVFCSGMCGWWLRQRR---KHAWGAFTFGLPVFPVVLTLAVLA
MAV_4686|M.avium_104 VLVASVVLGGAAAVWGAWQRRRG---SRPRAAFAFALPVLPVLLTLAILA
MSMEG_0904|M.smegmatis_MC2_155 VAALSFVGGAAAAWVGWRRYQQR---SRVWAAFLWCLPVSPVLFSLTLLG
TH_4134|M.thermoresistible__bu IAAVTAVTAAAAIWLGLRRYRQR---SHVWAAFLWSLPVLPVLFTLSVLA
Mflv_0113|M.gilvum_PYR-GCK VAAVSFVVGVSAALLGRRLWQRKDEKSEARAAFLWGLSISPVLLSLIVLG
Mvan_0795|M.vanbaalenii_PYR-1 VAGVSLILGLASALLGRRKLQRKDRKSEIWAAFLFGLSISPVLFSLAVLG
: : : . * : .** : *.: **:::: :*.
MMAR_0786|M.marinum_M DIYF
MUL_2215|M.ulcerans_Agy99 DIYF
Mb0472|M.bovis_AF2122/97 DIYL
Rv0463|M.tuberculosis_H37Rv DIYL
MLBr_02388|M.leprae_Br4923 KIYL
MAV_4686|M.avium_104 DVYL
MSMEG_0904|M.smegmatis_MC2_155 VTYL
TH_4134|M.thermoresistible__bu ATYL
Mflv_0113|M.gilvum_PYR-GCK VTYL
Mvan_0795|M.vanbaalenii_PYR-1 VTYL
*: