For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTYRLPDTKTVRDAAAHVRALVHPQIYNHSIRTYLLGAEAARRDGETDLDDEIFCVAALFHDSGTADEYN GPARFEIEGADAAAEFLSDRGFDADAVDAAWQAIALHTTPGIPERRGAIPHYLRTGVMIEFGPPELRQSY AEAIAAAEEDLPRHRLEQTLESLVVQQALANPHKAPRLSWAAELVAHHDPARDGISPGF
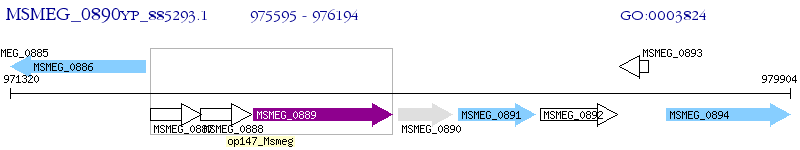
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0890 | - | - | 100% (199) | HD domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3487 | - | 1e-19 | 41.07% (112) | metal-dependent phosphohydrolase |
| M. smegmatis MC2 155 | MSMEG_5764 | - | 3e-05 | 29.75% (121) | putative cyanamide hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4753c | - | 8e-20 | 43.24% (111) | putative metal-dependent phosphohydrolase |
| M. marinum M | MMAR_1613 | - | 3e-25 | 36.72% (177) | hypothetical protein MMAR_1613 |
| M. avium 104 | MAV_2708 | - | 8e-20 | 33.51% (194) | metal-dependent phosphohydrolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3424 | - | 2e-25 | 36.72% (177) | hypothetical protein MUL_3424 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4753c|M.abscessus_ATCC_199 ------MTEIAGVEIPDTPLVREITEYIHDTEDDLLFHHSRRVYLFGVLQ
MAV_2708|M.avium_104 MTTTSSIETIADIAIPDTALVREITEFIRDAEDDLLFHHSRRVFLFGALQ
MMAR_1613|M.marinum_M --------MQSVPELVDTTVAQAALRLSRSTESPAVFNHSVRSYLFGELL
MUL_3424|M.ulcerans_Agy99 --------MQSVPELVDTTVAQAALRLSRSTESPAVFNHSVRSYLFGELL
MSMEG_0890|M.smegmatis_MC2_155 ----------MTYRLPDTKTVRDAAAHVRALVHPQIYNHSIRTYLLGAEA
: ** .: : :::** * :*:*
MAB_4753c|M.abscessus_ATCC_199 GQRRGVKP----DLELLYAGAMFHDIGLTDGYRGSPLRFEVDGANAARDF
MAV_2708|M.avium_104 GRRRGLQP----DLELLYAGAMFHDLGLTERYRDSALRFEVDGANAAREF
MMAR_1613|M.marinum_M AAHDGMRHGVDYDSQTLFLGCVLHDLGAGSAAPG-KARFEVEGADLAAAL
MUL_3424|M.ulcerans_Agy99 AAHDGMRHGVDYDSQTLFLGCVLHDLGAGSAAPG-KARFEVEGADLAAAL
MSMEG_0890|M.smegmatis_MC2_155 ARRDGE---TDLDDEIFCVAALFHDSGTADEYNG-PARFEIEGADAAAEF
. : * * : : ..::** * . . ***::**: * :
MAB_4753c|M.abscessus_ATCC_199 LIERGVGAEDAQNVWLAIALHTTPGITEFLSPEVALVTAGVETDVVGIDR
MAV_2708|M.avium_104 LLHRGVSKADADKVWLGIALHTTPGVPEFLDPEIALVTAGVETDVVGVGR
MMAR_1613|M.marinum_M LGEHGCDNEVVDTVWEAIALHTCFGIAERRGPICYLVHAGVGMDFG-RNA
MUL_3424|M.ulcerans_Agy99 LGEHGCDNEVVDTVWEAIALHTFFGIAERRGPICYLVHAGVGMDFG-RNA
MSMEG_0890|M.smegmatis_MC2_155 LSDRGFDADAVDAAWQAIALHTTPGIPERRGAIPHYLRTGVMIEFGPPEL
* .:* . .: .* .***** *:.* .. : :** :.
MAB_4753c|M.abscessus_ATCC_199 DALSGKDLDEVVAAHPRPDFKNRILRAFFEGNRHRPRSTFGNMNADVLAH
MAV_2708|M.avium_104 DDLAPEALAAVIAAHPRPDFKTRILQAFNDGMKHRPHSTFGTMNADVLEH
MMAR_1613|M.marinum_M EFVDDRTAALIHDQYPRLSMATTLVDAITTHAQRSPEAAPPYTVPGGLLL
MUL_3424|M.ulcerans_Agy99 EFVDDRTAALIHDQYPRLSMATTLVDAITTHAQRSPEAAPPYTVPGGLLL
MSMEG_0890|M.smegmatis_MC2_155 RQSYAEAIAAAEEDLPRHRLEQTLESLVVQQALANPHKAPRLSWAAELVA
. ** : : . *. : . *
MAB_4753c|M.abscessus_ATCC_199 FDPSFVRDDLVDLIHANSWPE
MAV_2708|M.avium_104 FDPTFARDNLVDLILNNGWPE
MMAR_1613|M.marinum_M ERRTNGITALEQLESLGRWGE
MUL_3424|M.ulcerans_Agy99 ERRTNGITALEQLESLGRWGE
MSMEG_0890|M.smegmatis_MC2_155 HHDPARDGISPGF--------
. :