For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RADSAPSTQALRGWQRKALVKYLTTKPRDYLLVATPGAGKTTFALRIAAELLADRTVDKITVVVPTEHLK IQWAASAARNGISLDPKFSNSNSQTSSEYDGVVVTYAQVASHPTRHRVRTENYRTLVIFDEIHHGGDAKS WGDAMREAYSDATRRLALTGTPFRSDDSPIPFVNYEPDESGFMRSQADHVYGYADALADGVVRPVVFLAY SGEARWRDSAGEEHAARLGEPLTAEQTARAWRTALNPAGEWMPAVIKAADRRLEQKRQHVPDAGGMIIAT NQTTARAYADLLTKITGETPTVVLSDDPSASDRISQYSESNSKWLVAVRMVSEGVDVPRLSVGVYATSAS TPLFFAQAIGRFVRSRRQGETASIFLPSVPNLLLLASEMEAQRNHVLGKPHRESDGLDDAALEEAEKRKD EKSELENGFEYLGADAELDQVIFDGASWGTATPAGSDEEADYLGIPGLLDAEQMRDLLRRRQEEQLTKRT AVAAETGAPPPPRTTHGQLRELRKELNALVTIAHHRTGKPHGWIHNELRRICGGPPVAAATTEQLKERIE AVRRLNP*
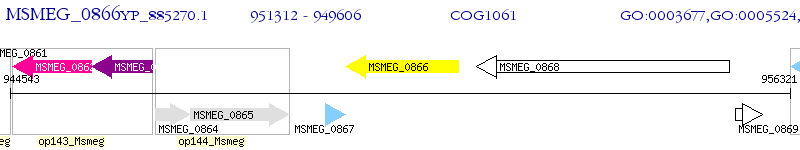
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0866 | - | - | 100% (568) | DNA or RNA helicase of superfamily protein II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2941 | - | 0.0 | 74.47% (568) | hypothetical protein Mb2941 |
| M. gilvum PYR-GCK | Mflv_0145 | - | 0.0 | 81.08% (576) | type III restriction enzyme, res subunit |
| M. tuberculosis H37Rv | Rv2917 | - | 0.0 | 74.30% (568) | hypothetical protein Rv2917 |
| M. leprae Br4923 | MLBr_01624 | - | 0.0 | 72.11% (563) | hypothetical protein MLBr_01624 |
| M. abscessus ATCC 19977 | MAB_0645c | - | 0.0 | 77.46% (568) | hypothetical protein MAB_0645c |
| M. marinum M | MMAR_1790 | - | 0.0 | 78.55% (564) | hypothetical protein MMAR_1790 |
| M. avium 104 | MAV_3772 | - | 0.0 | 76.24% (564) | DNA or RNA helicase of superfamily protein II |
| M. thermoresistible (build 8) | TH_1065 | - | 0.0 | 85.95% (477) | CONSERVED HYPOTHETICAL ALANINE AND ARGININE RICH PROTEIN |
| M. ulcerans Agy99 | MUL_2038 | - | 0.0 | 78.37% (564) | hypothetical protein MUL_2038 |
| M. vanbaalenii PYR-1 | Mvan_0759 | - | 0.0 | 82.57% (568) | type III restriction enzyme, res subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0866|M.smegmatis_MC2_155 ---------------------------MRADSA-----------------
TH_1065|M.thermoresistible__bu --------------------------------------------------
Mflv_0145|M.gilvum_PYR-GCK ---------------------------MRADTAPISDDA-----------
Mvan_0759|M.vanbaalenii_PYR-1 ---------------------------MRADAA-----------------
MLBr_01624|M.leprae_Br4923 -------MTQSDHRHTRPSSYGGTVLTQRVSQATVEDSR-----------
MAV_3772|M.avium_104 --------------MSVPVSS----LDPRAGNDHSDRSG-----------
Mb2941|M.bovis_AF2122/97 MRVTRLVDAESTRCDVGPAPKSVAMLHFTAATSRFRLGRERANSVRSDGG
Rv2917|M.tuberculosis_H37Rv MRVTRLVDAESTRCDVGPAPKSVAMLHFTAATSRFRLGRERANSVRSDGG
MMAR_1790|M.marinum_M ------------------------MQSVSPAEPIIEGTR-----------
MUL_2038|M.ulcerans_Agy99 ------------------------MQSVSPAEPIIEGTR-----------
MAB_0645c|M.abscessus_ATCC_199 ---------------------------MRAVEAPS---------------
MSMEG_0866|M.smegmatis_MC2_155 ---------PSTQALRGWQRKALVKYLTTKPRDYLLVATPGAGKTTFALR
TH_1065|M.thermoresistible__bu --------------LRGWQRKALVKYLTAKPRDFLAVATPGAGKTTFALR
Mflv_0145|M.gilvum_PYR-GCK -------GVPAVQALRSWQRRALVKYLTARPRDFLAVATPGAGKTTFALR
Mvan_0759|M.vanbaalenii_PYR-1 ---------PSTQALRGWQRRALVKYLSAAPRDFLAVATPGAGKTTFALR
MLBr_01624|M.leprae_Br4923 -------------PLRGWQRRAMVKYLASQPRDFLAVATPGSGKTTFALR
MAV_3772|M.avium_104 -------------ALRGWQRRALVKYLAGQPRDFLAVATPGSGKTTFALR
Mb2941|M.bovis_AF2122/97 WGVLQPVSATFNPPLRGWQRRALVQYLGTQPRDFLAVATPGSGKTSFALR
Rv2917|M.tuberculosis_H37Rv WGVLQPVSATFNPPLRGWQRRALVQYLGTQPRDFLAVATPGSGKTSFALR
MMAR_1790|M.marinum_M -------------PLRSWQRRALVKYLGTQPRDFLAVATPGSGKTAFALR
MUL_2038|M.ulcerans_Agy99 -------------PLRSWQRRALVKYLGTQPRDFLAVATPGSGKTVFALR
MAB_0645c|M.abscessus_ATCC_199 -----------THTLRGWQRRALVRYLAAKPRDFLAVATPGSGKTRFALR
**.***:*:*:** ***:* *****:*** ****
MSMEG_0866|M.smegmatis_MC2_155 IAAELLADRTVDKITVVVPTEHLKIQWAASAARNGISLDPKFSNSNSQTS
TH_1065|M.thermoresistible__bu IAAELLSDRTVDTVTVVVPTEHLKFQWAAAAESHGIALDPRFSNSDAQTS
Mflv_0145|M.gilvum_PYR-GCK IVSELLAEGTVDAVTIVVPTEHLKIQWAQAAARHGIALDPKFSNSNSQTS
Mvan_0759|M.vanbaalenii_PYR-1 IVAELLAEGTVEAVTIVVPTEHLKIQWAQAAARHGIALDPKFSNSNSQTS
MLBr_01624|M.leprae_Br4923 VMTELLNSHAVEQVTVVVPTEHLKVQWTRAAATHGLALDPKFSNTNPRTS
MAV_3772|M.avium_104 VAAELLGQRAVEQVTVVVPTEHLKVQWAQAAERHGLALDPRFSNSNPRIA
Mb2941|M.bovis_AF2122/97 IAAELLRYHTVEQVTVVVPTEHLKVQWAHAAAAHGLSLDPKFANSNPQTS
Rv2917|M.tuberculosis_H37Rv IAAELLRYHTVEQVTVVVPTEHLKVQWAHAAAAHGLSLDPKFANSNPQTS
MMAR_1790|M.marinum_M IAAEALGQRAAEQITVVVPTEHLKTQWAQAGARLGLSLDPKFSNSNAQTS
MUL_2038|M.ulcerans_Agy99 IAAEALGQRAAEQITVVVPTEHLKTQWAQAGARLGLSLDPKFSNSNAQTS
MAB_0645c|M.abscessus_ATCC_199 IATELLADGTVDKITVVVPTEHLKVQWAQSATSFGLSLDPRFSNSDSQTS
: :* * :.: :*:******** **: :. *::***:*:*::.: :
MSMEG_0866|M.smegmatis_MC2_155 SEYDGVVVTYAQVASHPTRHRVRTENYRTLVIFDEIHHGGDAKSWGDAMR
TH_1065|M.thermoresistible__bu SEYHGVVITYAQVASHPTRHRVRTENRRTLVIFDEIHHGGDNKSWGEAML
Mflv_0145|M.gilvum_PYR-GCK SEYHGVVVTYAQVASHPSRHRVRTENRKTLVVFDEIHHGGDAKSWGDAIR
Mvan_0759|M.vanbaalenii_PYR-1 SDYHGVVVTYAQVASHPTRHRVRTENRKTLVVFDEIHHGGDAKSWGDAIR
MLBr_01624|M.leprae_Br4923 PEYHGVTMTYAQVAAHPTLHRVRTEGRRTLVIFDEIHHGGNAKAWGDAIR
MAV_3772|M.avium_104 PEYHGVMVTYAQVAAHPTLHRVRTEQRRTLVIFDEIHHGGDAKTWGDAIR
Mb2941|M.bovis_AF2122/97 PEYHGVMVTYAQVASHPTLHRVRTEARKTLVVFDEIHHGGDAKTWGDAIR
Rv2917|M.tuberculosis_H37Rv PEYHGVMVTYAQVASHPTLHRVRTEARKTLVVFDEIHHGGDAKTWGDAIR
MMAR_1790|M.marinum_M AEYHGVVVTYAQVAAHPTLHRVRTEQRKTLVIFDEIHHGGDAKTWGDAIR
MUL_2038|M.ulcerans_Agy99 AEYHGVVVTYAQVAAHPTLHRVRTEQRKTLVIFDEIHHGGDAKTWGDAIR
MAB_0645c|M.abscessus_ATCC_199 SEYHGVVVTYAQVASHPTRHRVRTENHRTLVIFDEIHHAGDAKSWGEAVR
.:*.** :******:**: ****** :***:******.*: *:**:*:
MSMEG_0866|M.smegmatis_MC2_155 EAYSDATRRLALTGTPFRSDDSPIPFVNYEPDESGFMRSQADHVYGYADA
TH_1065|M.thermoresistible__bu EAFSDATRRLALTGTPFRSDDSPIPFVTYEPDENGFLRSRADYVYGYADA
Mflv_0145|M.gilvum_PYR-GCK EAFDDATRRLALTGTPFRSDDSAIPFVTYETGPDGFQRSQADHTYGYADA
Mvan_0759|M.vanbaalenii_PYR-1 EAFDDATRRLALTGTPFRSDDSPIPFVNYETGPDGFARSKADHTYGYSDA
MLBr_01624|M.leprae_Br4923 EAFSDATRRLALTGTPFRSDDKPIPFVTYALDADGLMHSQADHTYSYAEG
MAV_3772|M.avium_104 EAFGDATRRLALTGTPFRSDDSPIPFVRYEAGPDGVRRSQADHTYGYPEA
Mb2941|M.bovis_AF2122/97 EAFGDATRRLALTGTPFRSDDSPIPFVSYQPDADGVLRSQADHTYGYAEA
Rv2917|M.tuberculosis_H37Rv EAFGDATRRLALTGTPFRSDDSPIPFVSYQPDADGVLRSQADHTYGYAEA
MMAR_1790|M.marinum_M EAFSDASRRLALTGTPFRSDDSPIPFVNYEPDGEGVLRSQADHTYGYAEA
MUL_2038|M.ulcerans_Agy99 EAFSDASRRLALTGTPFRSDDSPIPFVNYEPDGEGVLRSQADHTYGYAEA
MAB_0645c|M.abscessus_ATCC_199 EAFDDATRRLSLTGTPFRSDDSPIPFINYEPDGAGYQRSQADHTYGYSEA
**:.**:***:**********..***: * . * :*:**:.*.*.:.
MSMEG_0866|M.smegmatis_MC2_155 LADGVVRPVVFLAYSGEARWRDSAGEEHAARLGEPLTAEQTARAWRTALN
TH_1065|M.thermoresistible__bu LADGVVRPVVFLAYSGEARWRDSAGEEYSARLGEPLSAEQTARAWRTALN
Mflv_0145|M.gilvum_PYR-GCK LADGVVRPVMFMAYSGEARWRDSAGEEHAARLGEPLTAEQTARAWRAALD
Mvan_0759|M.vanbaalenii_PYR-1 LADGVVRPVMFMAYSGEARWRDSAGEEHAARLGEPLTAEQTARAWKTALD
MLBr_01624|M.leprae_Br4923 LADGVVRPVVFLVYSGQARWRNSAGEEHAARLGEPLSAEQTARAWRTALD
MAV_3772|M.avium_104 LADGVVRPVVFLAYSGEARWRDSAGEEHAARLGEPLSAEQTARAWRTALD
Mb2941|M.bovis_AF2122/97 LADGVVRPVVFLAYSGQARWRDSAGEEYEARLGEPLSAEQTARAWRTALD
Rv2917|M.tuberculosis_H37Rv LADGVVRPVVFLAYSGQARWRDSAGEEYEARLGEPLSAEQTARAWRTALD
MMAR_1790|M.marinum_M LADGVVRPVVFLAYSGQARWRDSAGEEHEARLGEPLSAEQTARAWRTALD
MUL_2038|M.ulcerans_Agy99 LADGVVRPVVFLAYSGQARWRDSAGEEHEARLGEPLSAEQTARAWRTALD
MAB_0645c|M.abscessus_ATCC_199 LADGVVRPVVFMAYSGEARWRDSAGEEHAARLGEPLSAEQTARAWRTTLD
*********:*:.***:****:*****: *******:********:::*:
MSMEG_0866|M.smegmatis_MC2_155 PAGEWMPAVIKAADRRLEQKRQ-HVPDAGGMIIATNQTTARAYADLLTKI
TH_1065|M.thermoresistible__bu PAGDWMPAVIAAADKRLQQKRR-HVPDAGGMIIASDQTSARAYAKLLHQI
Mflv_0145|M.gilvum_PYR-GCK PKGEWMPAVIAAADKRLQGKRQ-HVPDAGGMIIATDQTAARAYAELLLKI
Mvan_0759|M.vanbaalenii_PYR-1 PKGEWMPAVIAAADKRLQGLRQ-HVPDAGGMIIASDQTTARAYADLLVKI
MLBr_01624|M.leprae_Br4923 PSGEWMPAVISAADQRLRQLRT-HVPDAGGMIIASDQTAARAYANLLAQM
MAV_3772|M.avium_104 PAGEWMPAVIAAADQRLRQLRA-HIPDAGGMIIASDRVAARAYATLLTEI
Mb2941|M.bovis_AF2122/97 PEGEWMPAVITAADRRLRQLRA-HVPDAGGMIIASDRTTARAYARLLTTM
Rv2917|M.tuberculosis_H37Rv PEGEWMPAVITAADRRLRQLRA-HVPDAGGMIIASDRTTARAYARLLTTM
MMAR_1790|M.marinum_M PAGEWMPAVIAAADRRLRQLRE-HVPDAGGMIIASDRTAARAYAALLQKM
MUL_2038|M.ulcerans_Agy99 PAGEWMPAVIAAADRRLRQLRE-HVPDAGGMIIASDRTAARAYAALLQKM
MAB_0645c|M.abscessus_ATCC_199 PNGEWIPAVLRAAHARLLQKRKGGMTDAGAMIIATDQKAARAYAKLLTQI
* *:*:***: **. ** * :.***.****::: :***** ** :
MSMEG_0866|M.smegmatis_MC2_155 TGETPTVVLSDDPSASDRISQYSESNSKWLVAVRMVSEGVDVPRLSVGVY
TH_1065|M.thermoresistible__bu TGEQPTVVLSDDPGASDRISEYSAGTSRWLVAVRMVSEGVDVPRLSVGVY
Mflv_0145|M.gilvum_PYR-GCK TGEAPTVVLSDDKGASDRISQFSAGTSRWLVAVRMVSEGVDVPRLSVGVY
Mvan_0759|M.vanbaalenii_PYR-1 TGEAPTVVLSDDKGASDRISEYSAGTSRWLVAVRMVSEGVDVPRLAVGVY
MLBr_01624|M.leprae_Br4923 TSETPTLVLSDDPGSSARITEFAKNTSQWLIAVRMVSEGVDIPRLSVGIY
MAV_3772|M.avium_104 TSETPTVVLSDDPGSSARISEFAASTSRWLVAVRMVSEGVDVPRLSVGIY
Mb2941|M.bovis_AF2122/97 TAEEPTVVLSDDPGSSARITEFAQGTSRWLVAVRMVSEGVDVPRLSVGVY
Rv2917|M.tuberculosis_H37Rv TAEEPTVVLSDDPGSSARITEFAQGTSRWLVAVRMVSEGVDVPRLSVGVY
MMAR_1790|M.marinum_M TGETPTVVLSDDPGSSERITEFSNGTSRWLVAVRMVSEGVDVPRLSVGIY
MUL_2038|M.ulcerans_Agy99 TGETPTVVLSDDPGSSERITEFSNGTSRWLVAVRMVSEGVDVPRLSVGIY
MAB_0645c|M.abscessus_ATCC_199 TGSEPTVVLSDDPKSSDRIAQFSASTSEWMVAVRMVSEGVDVPRLAVGVY
*.. **:***** :* **:::: ..*.*::**********:***:**:*
MSMEG_0866|M.smegmatis_MC2_155 ATSASTPLFFAQAIGRFVRSRRQGETASIFLPSVPNLLLLASEMEAQRNH
TH_1065|M.thermoresistible__bu ATSASTPLFFAQVIGRFVRSRRPGETASIFLPSVPNLLLLASELEAQRNH
Mflv_0145|M.gilvum_PYR-GCK ATSASTPLFFAQAIGRFVRSRRAGETASIFLPSVPNLLLLASEMEAQRNH
Mvan_0759|M.vanbaalenii_PYR-1 ATSASTPLFFAQAIGRFVRSRRPGETASIFLPSVPNLLLLASEMEAQRNH
MLBr_01624|M.leprae_Br4923 ATSASTPLFFAQAIGRFVRSRHPGETASIFVPSVPNLLQLASELETQRNH
MAV_3772|M.avium_104 ATSASTPLFFAQAVGRFVRSRRPGETASIFLPSVPNLLQLASELEAQRNH
Mb2941|M.bovis_AF2122/97 ATNASTPLFFAQAIGRFVRSRRPGETASIFVPSVPNLLQLASALEVQRNH
Rv2917|M.tuberculosis_H37Rv ATNASTPLFFAQAIGRFVRSRRPGETASIFVPSVPNLLQLASALEVQRNH
MMAR_1790|M.marinum_M ATSASTPLFFAQAIGRFVRSRRPGETASIFLPSVPNLLQLASELEAQRNH
MUL_2038|M.ulcerans_Agy99 ATSASTPLFFAQAIGRFVRSRRPGETASIFLPSVPNLLQLASELEAQRNH
MAB_0645c|M.abscessus_ATCC_199 ATSASTPLFFAQAIGRYVRSRTPGETASIFLPSVPTLLELASLLEEQRNH
**.*********.:**:**** *******:****.** *** :* ****
MSMEG_0866|M.smegmatis_MC2_155 VLGKPHRESDGLDDAALEEAE-KRKDEKSELENGFEYLGADAELDQVIFD
TH_1065|M.thermoresistible__bu VLGKPHRESDGLDDDLLRDAA-RTKDEKTELDNGYEALGAEAELDQLIFD
Mflv_0145|M.gilvum_PYR-GCK VLGKPHRET--LEDPLDAELAEQNRDEPGEEENKVEYLGADAELDQVIFD
Mvan_0759|M.vanbaalenii_PYR-1 VLGKPHREP--LEDPLDAELREQKRDEPGEEENKIEYLGADAELDQVIFD
MLBr_01624|M.leprae_Br4923 VLGKPHRES--TDNPLGGNPATMTQTEQDDTEKYFTAIGADAELDQIIFD
MAV_3772|M.avium_104 VLGEPHRVS--EGDPLDGDPATRTQNEKSELDNGFTSLGADAELDQVIFD
Mb2941|M.bovis_AF2122/97 VLGRPHRES--AHDPLDGDPATRTQTERGGAERGFTALGADAELDQVIFD
Rv2917|M.tuberculosis_H37Rv VLGRPHRES--AHDPLDGDPATRTQTERGGAERGFTALGADAELDQVIFD
MMAR_1790|M.marinum_M VLGKPHRES--LDDPLDGDPATKTQDEQGDLEKGFTSLGADAELDQVIFD
MUL_2038|M.ulcerans_Agy99 VLGKPHRES--LDDPLDGDPATKTQDEQGDLEKGFTSLGADAELDQVIFD
MAB_0645c|M.abscessus_ATCC_199 VLGKPHREP-----MGDEEPVERKRTEPGEEEKGFESLSASAELDQVIFD
***.*** . : : * :. :.*.*****:***
MSMEG_0866|M.smegmatis_MC2_155 GASWGTATPAGSDEEADYLGIPGLLDAEQMRDLLRRRQEEQLTKRTAVAA
TH_1065|M.thermoresistible__bu GASFGTATPAGSEEEADYLGIPGLLDAEQMRELLRRRQEEQLTX----XX
Mflv_0145|M.gilvum_PYR-GCK GSSFGTATPAGSDEEADYLGIPGLLDAASMRDLLRRRQDEQMQK-----R
Mvan_0759|M.vanbaalenii_PYR-1 GSSFGTATPAGSDEEADYLGIPGLLDADSMRDLLRRRQEEQLTK-----R
MLBr_01624|M.leprae_Br4923 GSSFGTATPAGSEEEAYYLGIPGLLDADQMRALLHRRQNEQLQKRTAAQQ
MAV_3772|M.avium_104 GSSFGTAAPAGSEEEADYLGIPGLLDAEQMRALLHQRQDQQLQRRAGQPS
Mb2941|M.bovis_AF2122/97 GSSFGTATPTGSDEEADYLGIPGLLDAEQMRALLHRRQDEQLRKRAQLQK
Rv2917|M.tuberculosis_H37Rv GSSFGTATPTGSDEEADYLGIPGLLDAEQMRALLHRRQDEQLRKRAQLQK
MMAR_1790|M.marinum_M GSSFGTATPAGSDEEADYLGIPGLLDADQMRALLHRRQDEQLQKRAQLQQ
MUL_2038|M.ulcerans_Agy99 GSSFGTATPAGSDEEADYLGIPGLLDADQMRASLHRRQDEQLQKRAQLQQ
MAB_0645c|M.abscessus_ATCC_199 GASFGTATPAGSEEEAEYLGIPGLLDPNQMRDLLRRRQEEQLTKR---TA
*:*:***:*:**:*** *********. .** *::**::*:
MSMEG_0866|M.smegmatis_MC2_155 ETGAPP----PPRTTHGQLRELRKELNALVTIAHHRTGKPHGWIHNELRR
TH_1065|M.thermoresistible__bu TASSAL----ALRTSRGNAA------------------------------
Mflv_0145|M.gilvum_PYR-GCK TESGVI----PPKTTHGQLRDLRSELNTLVSLAHHRTGRPHGWIHNELRR
Mvan_0759|M.vanbaalenii_PYR-1 TESGLA----VPKTTHGQLRDLRSELNTLVSLAHHRTGRPHGWIHNELRR
MLBr_01624|M.leprae_Br4923 ASSTPDRTSGAPASVHGQLRELRRELNSLVSIAHHHTGKPHGWIHNELRR
MAV_3772|M.avium_104 AGDAP------PATVHGQLRELRRELNTLVSIAHHRTGKPHGWIHNELRR
Mb2941|M.bovis_AF2122/97 GATQPA-TSGASASVHGQLRDLRRELHTLVSIAHHRTGKPHGWIHDELRR
Rv2917|M.tuberculosis_H37Rv GATQPA-TSGASASVHGQLRDLRRELHTLVSIAHHRTGKPHGWIHDERRR
MMAR_1790|M.marinum_M GVTQVG-TLAEPATTHGQIRELRRELNTLVSIAHHRTGKPHGWIHNELRR
MUL_2038|M.ulcerans_Agy99 GVTQVG-TLAEPATTHGQIRELRRELNTLVSIAHHRTGKPHGWIHNELRR
MAB_0645c|M.abscessus_ATCC_199 SGEVPP----EITSTHGQLRELRRELNALVSATHFKSGKPHGWIHNELRR
: :*:
MSMEG_0866|M.smegmatis_MC2_155 ICGGPPVAAATTEQLKERIEAVRRLNP----
TH_1065|M.thermoresistible__bu -------------------------------
Mflv_0145|M.gilvum_PYR-GCK RCGGPPVAAATREQLQERIEAVRVLQRELTA
Mvan_0759|M.vanbaalenii_PYR-1 RCGGPPVAAATREQLQERIEAVRVLQRELSA
MLBr_01624|M.leprae_Br4923 RCGGPPIAAATHDQLKARIDAVRQLNAEPS-
MAV_3772|M.avium_104 RCGGPPIAAASREQLRARIDAVRRLNAEHS-
Mb2941|M.bovis_AF2122/97 RCGGPPIAAATRAQIKARIDALRQLNSERS-
Rv2917|M.tuberculosis_H37Rv RCGGPPIAAATRAQIKARIDALRQLNSERS-
MMAR_1790|M.marinum_M RCGGPPIAAATRDQLKARIDAVRQLNAEHG-
MUL_2038|M.ulcerans_Agy99 RCGGPPIAAATRDQLKARIDAVRQLNAEHG-
MAB_0645c|M.abscessus_ATCC_199 RCGGPPVAAATRDQLKERIEAVRTLHREL--