For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NIRPLRQSVRPSPIFLLVIAVTAAGGALAWIAADTIRPLSYVGVFILVIAGWLMSLCLHEFGHAFTAWRF GDHGVETRGYLTLNPLKYTHPMLSLGLPVLIIALGGIGFPGGAVYLQTHWMTARQKSIVSLAGPAANLVL AVLLLGLTRAFWDPAHAVFWSGIAFLGFLQVTALVLNLLPIPGLDGYGALEPHLNPETQRALAPAKQWGF LIVVVLLITPALNRWFFELVYWFFDFSGVSSYLVSAGGQLTRFWSAWF*
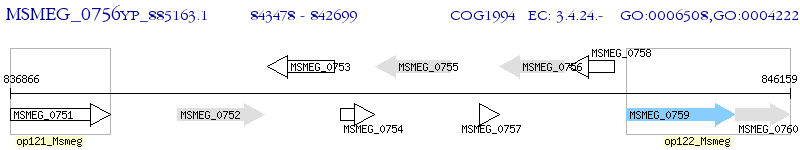
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0756 | - | - | 100% (259) | peptidase, M50B family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0366 | - | 1e-106 | 69.17% (253) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0230 | - | 1e-112 | 74.30% (249) | peptidase M50 |
| M. tuberculosis H37Rv | Rv0359 | - | 1e-106 | 68.77% (253) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4251 | - | 3e-92 | 62.55% (251) | hypothetical protein MAB_4251 |
| M. marinum M | MMAR_0678 | - | 1e-101 | 66.80% (253) | hypothetical protein MMAR_0678 |
| M. avium 104 | MAV_4781 | - | 1e-107 | 69.69% (254) | peptidase, M50 family protein |
| M. thermoresistible (build 8) | TH_0137 | - | 1e-107 | 67.44% (258) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_2836 | - | 5e-99 | 65.22% (253) | hypothetical protein MUL_2836 |
| M. vanbaalenii PYR-1 | Mvan_0674 | - | 1e-114 | 72.87% (258) | peptidase M50 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0230|M.gilvum_PYR-GCK --MVS-LVG-VRPSPVFAALVAVTAAGGALAWTSGDDVRPLSYAAVFVLV
Mvan_0674|M.vanbaalenii_PYR-1 MSVRP-LHQSVRPSPIFLALVAVTAAGGALAWTSAGRIEPVAYIGVFVFV
TH_0137|M.thermoresistible__bu VSIRS-VSGSVRPSPVFLAIVALTAAGGGLAWASATVTGPAAYAGVFLFV
MSMEG_0756|M.smegmatis_MC2_155 MNIRP-LRQSVRPSPIFLLVIAVTAAGGALAWIAADTIRPLSYVGVFILV
Mb0366|M.bovis_AF2122/97 MSETG-QRESVRPSPIFLGLLGLTAVGGALAWLAGETVQPLAYAGVFVMV
Rv0359|M.tuberculosis_H37Rv MSETG-QRESVRPSPIFLGLLGLTAVGGALAWLAGETVQPLAYAGVFVMV
MMAR_0678|M.marinum_M ---MG-LHESVRPSPIFLGLVGLTTVGAALAWLAGSSVRPLAYAGVFVFV
MUL_2836|M.ulcerans_Agy99 MSEMG-LHESVRPSPIFLGLVGLTTVGAALAWLTGSSVRPLAYAGVFVFA
MAV_4781|M.avium_104 MSFRAPQHESVRPSPIFLALLGLTALGGALAWLAGSSPRPLAYLGVFVFV
MAB_4251|M.abscessus_ATCC_1997 ---MN-VKQSVRPSPVFLVLVALTVAGAVFAWLGGDSTEPLARGGVFVFV
*****:* ::.:*. *. :** . * : .**::.
Mflv_0230|M.gilvum_PYR-GCK IFGWLVTLCLHEFGHAYSAYRFGDRDVAVRGYLTLNPLKYTHPLLSLGLP
Mvan_0674|M.vanbaalenii_PYR-1 IFGWLVTLCLHEFGHAYSAYRFGDRDVAVRGYLTLNPLKYSHPLLSLGLP
TH_0137|M.thermoresistible__bu IAGWLVTLCLHEFAHAWSAWRFGDREVAARGYLTLNPLRYTHPLLSIGLP
MSMEG_0756|M.smegmatis_MC2_155 IAGWLMSLCLHEFGHAFTAWRFGDHGVETRGYLTLNPLKYTHPMLSLGLP
Mb0366|M.bovis_AF2122/97 IAGWLVSLCLHEFGHAFTAWRFGDHDVAVRGYLTLDPRRYSHPMLSLGLP
Rv0359|M.tuberculosis_H37Rv IAGWLVSLCLHEFGHAFTAWRFGDHDVAVRGYLTLDPRRYSHPMLSLGLP
MMAR_0678|M.marinum_M IAGWLVSLCLHEFGHAFTAWRFGDHDVAVRGYLTLDPRRYSHPALSLLLP
MUL_2836|M.ulcerans_Agy99 IAGWLVSLCLHEFGHAFTAWRFGDHDVAVRGYLTLDPRRYSHPALSLLLP
MAV_4781|M.avium_104 IAGWLVSLCLHEFGHAVTAWRFGDRDAAVRGYLTLDPRRYSHPALSLVLP
MAB_4251|M.abscessus_ATCC_1997 FAGWIVSLCLHEFGHAYTAWRFG--DTQARGYLTLNPLQYTDPVLSIALP
: **:::******.** :*:*** . .******:* :*:.* **: **
Mflv_0230|M.gilvum_PYR-GCK VLFIALGGIGLPGGAVYVQTALMTDRQKTVVSLAGPAVNLLFAVLLLGLT
Mvan_0674|M.vanbaalenii_PYR-1 VLFIALGGIGLPGGAVYVQTAHMTDRQKTLVSLAGPGVNLIFAVLLLALT
TH_0137|M.thermoresistible__bu VLFVALGGIGFPGGAVDVRTHRMTPWRQSVVSLAGPSVNLVFAALLLTLT
MSMEG_0756|M.smegmatis_MC2_155 VLIIALGGIGFPGGAVYLQTHWMTARQKSIVSLAGPAANLVLAVLLLGLT
Mb0366|M.bovis_AF2122/97 MLFIALGGIGLPGAAVYVHTWFMTTARRTLVSLAGPTVNLALAMLLLAAT
Rv0359|M.tuberculosis_H37Rv MLFIALGGIGLPGAAVYVHTWFMTTARRTLVSLAGPTVNLALAMLLLAAT
MMAR_0678|M.marinum_M MVFIALGGIGLPGAAVYVRTWFMTPTRRTLVSLAGPAANVVLAVLLLTAT
MUL_2836|M.ulcerans_Agy99 MVFIALGGIGLPGAAVYVRTWFMTPTRRTLVSLAGPAANVVLAVLLLTAT
MAV_4781|M.avium_104 MVIIALGGIGLPGAAVYVQTWFMTPARRTLVSLAGPAANLVLAAVLLALT
MAB_4251|M.abscessus_ATCC_1997 LVFIALGGIGLPGGAVYVHTAGMTNRQRTIVSLAGPTANAILAVLILGTT
::::******:**.** ::* ** ::::****** .* :* ::* *
Mflv_0230|M.gilvum_PYR-GCK RPMYDPAHAVFWAGVAFLGFLQVTAFVLNMLPIPGLDGYGALEPHLSPET
Mvan_0674|M.vanbaalenii_PYR-1 RLLYDPAHSVFWAGVAFLGFLQVTALLLNMLPIPGLDGYGALEPHLSPDT
TH_0137|M.thermoresistible__bu RLCYDPAHGVFWAGVAFLGFLQITALVLNLLPIPGLDGFGALEPHLSPPT
MSMEG_0756|M.smegmatis_MC2_155 RAFWDPAHAVFWSGIAFLGFLQVTALVLNLLPIPGLDGYGALEPHLNPET
Mb0366|M.bovis_AF2122/97 RLLFDPIHAVLWAGVAFLAFLQLTALVLNLLPIPGLDGYAALEPHLRPET
Rv0359|M.tuberculosis_H37Rv RLLFDPIHAVLWAGVAFLAFLQLTALVLNLLPIPGLDGYAALEPHLRPET
MMAR_0678|M.marinum_M RLFYDQDHWVLWAGVAFLGFLQIMAVVLNLLPIPGLDGYDALEPHLSPQT
MUL_2836|M.ulcerans_Agy99 RLFYDQDHWVLWAGVAFLGFLQIMAVVLKLLPIPGLDGYDALEPHLSPQT
MAV_4781|M.avium_104 RAFFTADHAVLWAGVAFLGFLQLTAVLLNLLPIPGLDGYDALEPHLSPET
MAB_4251|M.abscessus_ATCC_1997 ALFADREHVVFWAAMAYLGFLQVTAVVLNLLPVPGLDGYGALEPHLSPNT
* *:*:.:*:*.***: *.:*::**:*****: ****** * *
Mflv_0230|M.gilvum_PYR-GCK RRALAPARQWGFFILLILLIAPPLNQWFFAAVYWLFELSGVDVYLSQLGG
Mvan_0674|M.vanbaalenii_PYR-1 QRALNPAKQWGFFILLILLIAPPLNQWFFGAVYWLFELSGVPPALSAIGG
TH_0137|M.thermoresistible__bu RRAVQPAKQWGFLILIVLLLTPTLNQWFFSAVHWLYELSGAPGALSAAGS
MSMEG_0756|M.smegmatis_MC2_155 QRALAPAKQWGFLIVVVLLITPALNRWFFELVYWFFDFSGVSSYLVSAGG
Mb0366|M.bovis_AF2122/97 QRALAPAKQFALVFLLVLFLAPTLNGWFFGVVYWLFDLSGVSHRLAAAGS
Rv0359|M.tuberculosis_H37Rv QRALAPAKQFALVFLLVLFLAPTLNGWFFGVVYWLFDLSGVSHRLAAAGS
MMAR_0678|M.marinum_M QRALAPAKQFGIFILLFLLLAPVLNQWLFEFVGWVFDFSGVPHLLAMVGN
MUL_2836|M.ulcerans_Agy99 QRALAPAKQFGIFILLFLLLAPVLNQWLFEFVGWVSDFSGVPHLLAMVGN
MAV_4781|M.avium_104 QRALAPAKQWGFFILLFLLLAPGLNRWFFGIVYWLFDFSGVPHWLAGAGN
MAB_4251|M.abscessus_ATCC_1997 QRTLGNAKTFGFLILFALLMSPPVNRAFFGAVYWIVAYLGVDPALAQIGG
:*:: *: :.:.::. *:::* :* :* * *. *. * *.
Mflv_0230|M.gilvum_PYR-GCK GLTRFWSPWL
Mvan_0674|M.vanbaalenii_PYR-1 QLTRFWSAWT
TH_0137|M.thermoresistible__bu RLTRFWSSWM
MSMEG_0756|M.smegmatis_MC2_155 QLTRFWSAWF
Mb0366|M.bovis_AF2122/97 VLTRFWSIWF
Rv0359|M.tuberculosis_H37Rv VLARFWSIWF
MMAR_0678|M.marinum_M SLTRFWSQWI
MUL_2836|M.ulcerans_Agy99 SLTRFWSQWI
MAV_4781|M.avium_104 VLTRFWSRWV
MAB_4251|M.abscessus_ATCC_1997 WLFRFWSKLL
* ****