For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEPHARGRFAMTGTDGRAAVLGRIRSALAAVPPQPVTVPREYHREPLSGPADVERFAEAVAEYRARVHRV AASDIAGLVRELTGSDATVVVPPDVPAEWITGVRILSDDPVLTVEQLDAADAVLTGCAVGIAATGTIVLD AGATQGRRALTLVPDHHICVVRTDQIVDIVPQGFAALGTERPLTFISGPSATSDIELERVEGVHGPRTLD VLIL*
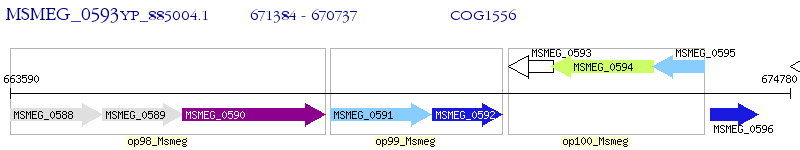
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0593 | - | - | 100% (215) | hypothetical protein MSMEG_0593 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3702 | - | 5e-43 | 51.94% (206) | hypothetical protein Mflv_3702 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2711 | - | 2e-42 | 49.51% (206) | hypothetical protein Mvan_2711 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3702|M.gilvum_PYR-GCK -----------MS--EAREQILGRIRAALSDRPDAP-GVPWTYGSAVGTG
Mvan_2711|M.vanbaalenii_PYR-1 -----------MS--EARAQILARIRAALGDRPAVP-EVPWTYGRPVGTG
MSMEG_0593|M.smegmatis_MC2_155 MVEPHARGRFAMTGTDGRAAVLGRIRSALAAVPPQPVTVPREYHREPLSG
*: :.* :*.***:**. * * ** * :*
Mflv_3702|M.gilvum_PYR-GCK AVDVVDRFIERVADYRAQVSRVDDAGP--AIALALNGFGHVVADEDVIRE
Mvan_2711|M.vanbaalenii_PYR-1 GLDLVDRFVERVADYRATIERVGEAGVGEAIALALRGLRKVVADGVVREA
MSMEG_0593|M.smegmatis_MC2_155 PAD-VERFAEAVAEYRARVHRVAASDIAGLVRELTGSDATVVVPPDVPAE
* *:** * **:*** : ** :. : . **. *
Mflv_3702|M.gilvum_PYR-GCK WPEVESWVSDS-GLSAAELDAVDAVVTTATVGIANTGTIVLSHGPGQGRR
Mvan_2711|M.vanbaalenii_PYR-1 WPDSADWVGDD-GLTASQLDGVDAVVTTATVAVANTGTIVLSHGPGQGRR
MSMEG_0593|M.smegmatis_MC2_155 WITGVRILSDDPVLTVEQLDAADAVLTGCAVGIAATGTIVLDAGATQGRR
* :.*. *:. :**..***:* .:*.:* ******. *. ****
Mflv_3702|M.gilvum_PYR-GCK ALSLVPDVHVCVVRVDQIVDDVPHAVARLIESGLHTRPLTWISGPSATSD
Mvan_2711|M.vanbaalenii_PYR-1 ALSLVPDVHVCVVRADQIVDDMPKAVARLMDSELHTRPLTWISGPSATSD
MSMEG_0593|M.smegmatis_MC2_155 ALTLVPDHHICVVRTDQIVDIVPQGFAALGT----ERPLTFISGPSATSD
**:**** *:****.***** :*:..* * ****:*********
Mflv_3702|M.gilvum_PYR-GCK IELDRVEGVHGPRTLRVIVVG
Mvan_2711|M.vanbaalenii_PYR-1 IELDRVEGVHGPRTLHVIVVG
MSMEG_0593|M.smegmatis_MC2_155 IELERVEGVHGPRTLDVLIL-
***:*********** *:::