For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAINVHPALDNLAIELPSWAFGNSGTRFKVFGTPGTPRTVEEKIADAAMVHRMTGLAPRVALHIPWDTV DDYAALGVYAREQGVELGTINSNTFQDDDYKFGSLTHTDKGVRQKAIDHHLACIEIMNQTGSRDLKIWLA DGTNYPGQGDIRGRQDRLAESLAAIYQHIGPDQRMVLEYKFFEPAMYMTDVPDWGTAYAQVSALGERAVV CLDTGHHAPGTNIEFIVAQLLRLGKLGSFDFNSRFYADDDLIVGAADPFQLFRILFEVIRGGGLDPSSGV ALMLDQCHNVEAKIPGQIRSVLNVAEMTARALLVDTEALAEAQENGDVLGANGIFMDAFYTDVRPALAKW RVDRGLPGDPMAAYAASGYQDTINAERVGGTQSSWGA
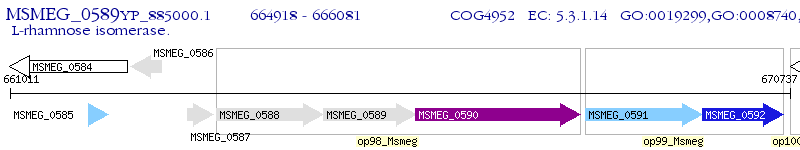
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0589 | rhaI | - | 100% (387) | L-rhamnose isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2175 | - | 8e-05 | 31.45% (124) | hypothetical protein MAB_2175 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0589|M.smegmatis_MC2_155 ----------MSAINVHPALDNLAIELPSWAFGNSGTRFKVFG-TPGTPR
MAB_2175|M.abscessus_ATCC_1997 MTQTVRPDEWHSDKWSHESIMDMQARMAPDDIRRIADEWKVTLRALDALL
* * :: :: .:.. : . . .:** : .:
MSMEG_0589|M.smegmatis_MC2_155 TVEEKIADAAMVHRMTGLAPRVALHIPWDTVDD-----------YAALGV
MAB_2175|M.abscessus_ATCC_1997 AGFLEDVTATIGDGWSGPGARAALSTMREYVENSRTALDTAFTLSSGLDV
: : . *:: . :* ..*.** : *:: :.*.*
MSMEG_0589|M.smegmatis_MC2_155 YAREQGVELGTINSNTFQDDDYKFGSLTHTDKGVRQKAIDHHLACIEIMN
MAB_2175|M.abscessus_ATCC_1997 LAQATSELQQKIAPPPVPVPAYKLGEIPRWGTPFAEQLALWEQAVNQVET
*: . .* . .. **:*.:.: .. . :: . * :: .
MSMEG_0589|M.smegmatis_MC2_155 QTGSRDLKIWLADGTNYPGQGDIRGRQDRLAESLAAIYQHIGPDQRMVLE
MAB_2175|M.abscessus_ATCC_1997 VYSAPAIRAAHAVAELIGPRESLRFGSGPDADVAVAGRRIPDEQAERAEE
.: :: * . : .:* .. *: .* : . : . . *
MSMEG_0589|M.smegmatis_MC2_155 YKFFEPAMYMTDVPDWGTAYAQVSALGER-AVVCLDTGHHAPGTNIEFIV
MAB_2175|M.abscessus_ATCC_1997 FLARWGLGESEGNPPQGTSPNNVGTPPQIPVMVPVMAGNPFSGKGFGIGS
: . * **: :*.: : .:* : :*: .*..: :
MSMEG_0589|M.smegmatis_MC2_155 AQLLRLGKLGSFDFNSRFYADDDLIVGAADPFQLFRILFEVIRGGGLDPS
MAB_2175|M.abscessus_ATCC_1997 APGSRGDELG----EDDVYASQNWMRDPLGRENPTRSAGIESAVPGLRGA
* * .:** :. .**.:: : .. . : * ** :
MSMEG_0589|M.smegmatis_MC2_155 SGVALMLDQCHNVEAKIPGQIRSVLNVAEMTARALLVDTEALAEAQENGD
MAB_2175|M.abscessus_ATCC_1997 LSPDRIPGFGTGAHVAGPGGIGGAAANRSWGAMMPMMGTYPAHAGQRRGD
. : . .... ** * .. . * ::.* . .*..**
MSMEG_0589|M.smegmatis_MC2_155 VLGANGIFMDAFYTDVRPALAKWRVDRGLPGDPMAAYAASGYQDTINAER
MAB_2175|M.abscessus_ATCC_1997 ENEHYSPPYLVNADNTRELLGDLPKG----WAPVIGLWESDSDEEFGPTP
. . :.* *.. . *: . *. :: :..
MSMEG_0589|M.smegmatis_MC2_155 VGGTQSSWGA
MAB_2175|M.abscessus_ATCC_1997 RRGFRSR---
* :*