For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MALIPMPHVDVADEQVAAVLSRAVGPIGLILDAVADLDPLGLRRRTHYLGEGDGVVGTVLDRIACVLDCA DIPGTRSWEGKDRAERIGWWVHGVGALDTVVVAFPGVFGVVADRLPLQDLLGFTNQALVLCAVAREYGVT DHDTQVRMLAAVLCGRDLAQTPQAQTEDESGDMSLVQRVWHLAGILNAIGDELGRRPHPRAPFRYLGMLP WVGAVADYLGEYGALVRAADAGQKWIAQHGAAADDRAPLRV
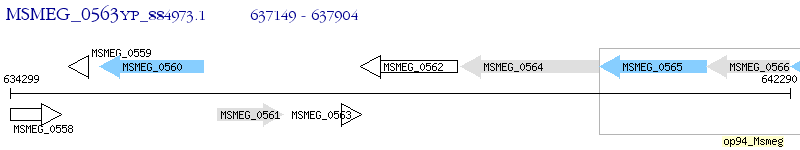
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0563 | - | - | 100% (251) | hypothetical protein MSMEG_0563 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0306 | - | 1e-60 | 50.00% (244) | hypothetical protein Mflv_0306 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3492c | - | 6e-69 | 52.65% (245) | hypothetical protein MAB_3492c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0440 | - | 5e-63 | 51.67% (240) | hypothetical protein Mvan_0440 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0306|M.gilvum_PYR-GCK -----------------------MNLLLSALPAPAVHITDTGVEAALSAA
Mvan_0440|M.vanbaalenii_PYR-1 ---MGSAG-----------HEKTVSPLPGMLPTPDVQITDAGVDATLAAA
MSMEG_0563|M.smegmatis_MC2_155 ---------------------------MALIPMPHVDVADEQVAAVLSRA
MAB_3492c|M.abscessus_ATCC_199 MERVKSAGSVCTVRALALCRRLGDDLFMGLISLPTAVLSDADIADALSRA
. :. * . ::* : .*: *
Mflv_0306|M.gilvum_PYR-GCK VRIINPLLDVLWETDPLDLKRRD--------------DALGRLLNAVDVP
Mvan_0440|M.vanbaalenii_PYR-1 VAIINPLLDVLWGTDPLGLKRRD--------------DGVGWALNAADLP
MSMEG_0563|M.smegmatis_MC2_155 VGPIGLILDAVADLDPLGLRRRTHYLGEGDGVVGTVLDRIACVLDCADIP
MAB_3492c|M.abscessus_ATCC_199 VRVINPVLTVLAQSDPFGLKERTYELGAGEGVVDKALDALACALNTATTP
* *. :* .: **:.*:.* * :. *: . *
Mflv_0306|M.gilvum_PYR-GCK GTPSWEDMDTDARIHWWVWRVGAVNTVAVAFPGVLGIVARQLPLQDLFGF
Mvan_0440|M.vanbaalenii_PYR-1 GTPAWDDMDADARIHWWVWRVGALNTVAVAFPGVLGILARRLPIQDLLGF
MSMEG_0563|M.smegmatis_MC2_155 GTRSWEGKDRAERIGWWVHGVGALDTVVVAFPGVFGVVADRLPLQDLLGF
MAB_3492c|M.abscessus_ATCC_199 GTKAWDKLDTRGKTHWWVQRVGAVNTVFVAFPGIFGALAARLPVQDLLGF
** :*: * : *** ***::** *****::* :* :**:***:**
Mflv_0306|M.gilvum_PYR-GCK VSQAVVLCAVARELGVTDQRTQVRMLAAVLCERDLSVVVFDGDDRRPLAS
Mvan_0440|M.vanbaalenii_PYR-1 VSQAIVLCAVARELGVTDQRPQVRMLAAVLCDRDLSVVVYQPSTDRRLVD
MSMEG_0563|M.smegmatis_MC2_155 TNQALVLCAVAREYGVTDHDTQVRMLAAVLCGRDLAQTPQAQTEDESGD-
MAB_3492c|M.abscessus_ATCC_199 SNQAIVLVAIARELGINDHREQVRLLAKVLCNRTLGDEALVGADDQAPET
.**:** *:*** *:.*: ***:** *** * *. .
Mflv_0306|M.gilvum_PYR-GCK IP-HSATGVAKAIWNLVGLFDAIADELAKRPHPRGPIKYLGLLPGVGAVA
Mvan_0440|M.vanbaalenii_PYR-1 IP-HHPLGIVSAVWNLVGLFDAIGDEVGKRPQPRGPFRYLGLLPAVGAVA
MSMEG_0563|M.smegmatis_MC2_155 ------MSLVQRVWHLAGILNAIGDELGRRPHPRAPFRYLGMLPWVGAVA
MAB_3492c|M.abscessus_ATCC_199 PRNWSPFALARTLWHLAGILRAIGDELEKRPQPKPIFRYLGMLPAVGSVA
.:. :*:*.*:: **.**: :**:*: ::***:** **:**
Mflv_0306|M.gilvum_PYR-GCK AYIGECGALARAAKQGRRWLEKP---------ARSEP
Mvan_0440|M.vanbaalenii_PYR-1 AYFGECGALARAAKMGRRWIERQ---------SQT--
MSMEG_0563|M.smegmatis_MC2_155 DYLGEYGALVRAADAGQKWIAQHGAAADDRAPLRV--
MAB_3492c|M.abscessus_ATCC_199 GYIGEYGALRRAASAGERSIAQS-------RPVRT--
*:** *** ***. *.: : : :