For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEQRTEEQSGQSGQPRQSGQSALGYAVAGVLVVIALGAATKFLETQVPQWFAGTSFAKAAKSIEFPVYAI ALGLIGNAILSKLALRDALSRGFRTEFFIKTGLVLLGASINLKVLVTAAGPAIIQALLLISIVFGFTWWL GGRLGLEDKLRALLASAVSICGVSAAIAAAGAVQAKKEQLAYAASLVIVFALPSIFLLPWLADVFGLSDA VAGAWIGGNIDTTAAVAASGAIAGEDALQIATIVKTTQNALIGFVAIALTAYFALKVERRTAADARPSLS QFWDRFPKFVLGFIAASIIGTLYLQFAADGKAAIATVNDLRNWFLIFAFVAIGLEFSARGLREAGWRPIA VFASATVVNIVVALGLATVLFGSFTLD*
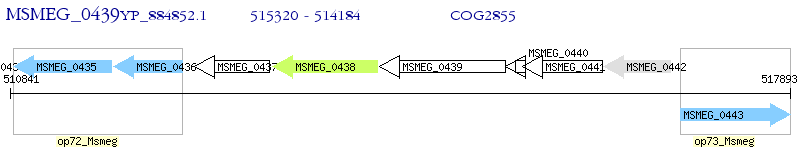
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0439 | - | - | 100% (378) | hypothetical protein MSMEG_0439 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3433c | - | 5e-50 | 34.14% (372) | hypothetical protein MAB_3433c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2056 | - | 6e-45 | 32.75% (345) | hypothetical protein MAV_2056 |
| M. thermoresistible (build 8) | TH_0929 | - | 3e-57 | 79.26% (135) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0439|M.smegmatis_MC2_155 ---MTEQRTEEQSGQSGQPRQSGQSALGYAVAGVLVVIALGAATKFLETQ
TH_0929|M.thermoresistible__bu ---------------------------------------VGAATRYLEAR
MAB_3433c|M.abscessus_ATCC_199 MTSKAEAHTA-DEAFENPEESFTSPRPLDYVPGILLLIGVGLLGKYAQIW
MAV_2056|M.avium_104 MTGVSTVRVAPDETDETADATFTSTRPLDYLPGILLLIGVGVLGKYAQIW
:* :: :
MSMEG_0439|M.smegmatis_MC2_155 VPQWFAGTSFAKAAKSIEFPVYAIALGLIGNAILSKLALRDALSRGFRT-
TH_0929|M.thermoresistible__bu VPDWADGTPFSRIADSIEFPVYAIALGLLGNVVLTRLAVREALAGGFRT-
MAB_3433c|M.abscessus_ATCC_199 WKDLAKAQHWT--VPDIEYVLWAIVIGLL---ITNTVGLHRIFRPGVQTY
MAV_2056|M.avium_104 WNALAKHEHWT--VPDIEYVLWAIVIGLV---ITNTVGLHPIFRPGVLTY
:: . .**: ::**.:**: : . :.:: : *. *
MSMEG_0439|M.smegmatis_MC2_155 EFFIKTGLVLLGASINLKVLVTAAGPAIIQALLLISIVFGFTWWLGGRLG
TH_0929|M.thermoresistible__bu EFFIKTGLMLLGASVNLKVLVTAAGPAIVQALLLISIVFGFTWWLGGRVG
MAB_3433c|M.abscessus_ATCC_199 EFWLKIGIVALGARFVLGDVLKLGGTSLVQILVDMTVAGAIILLAAKWFG
MAV_2056|M.avium_104 EFWLKAGIVALGSRFVLGDIAKLGGISLVQILVDMTIAGTIIIAVARWFG
**::* *:: **: . * : . .* :::* *: :::. : . .*
MSMEG_0439|M.smegmatis_MC2_155 LEDKLRALLASAVSICGVSAAIAAAGAVQAKKEQLAYAASLVIVFALPSI
TH_0929|M.thermoresistible__bu LDAKLRALLASAVSICGVSAAIAAA-------------------------
MAB_3433c|M.abscessus_ATCC_199 LSGKLGSLLAIGTSICGVSAIIAAKGAIRARNSDVSYAIASILALGAVAL
MAV_2056|M.avium_104 LSGKLGSLLAIGTSICGVSAIVAAKGAIRARNSDVSYAIAAILALGAVSL
*. ** :*** ..******* :**
MSMEG_0439|M.smegmatis_MC2_155 FLLPWLADVFGLSDAVAGAWIGGNIDTTAAVAASGAIAGEDALQIATIVK
TH_0929|M.thermoresistible__bu ------------------------------------------------VK
MAB_3433c|M.abscessus_ATCC_199 FTLPAIGHGLGLSDYEFGLWAGLAVDNTAETVATGRLYSEHAGDIATVVK
MAV_2056|M.avium_104 FVLPPLGHAIGLTDHEFGLWAGLSVDNTAETTATGYLYSEHAGKIAVLVK
**
MSMEG_0439|M.smegmatis_MC2_155 TTQNALIGFVAIALTAYFALKVERRTAAD-ARPSLSQFWDRFPKFVLGFI
TH_0929|M.thermoresistible__bu TTRTR---------------------------------------------
MAB_3433c|M.abscessus_ATCC_199 STRNALIGFVVLGFALYWAARGEAETLAPGIKAKAAFVWDKFPKFVLGFL
MAV_2056|M.avium_104 STRNALIGFVVLGFALFWAGRGQADEIAPGVRAKAAFIWAKFPKFVLGFL
:*:.
MSMEG_0439|M.smegmatis_MC2_155 AASIIGTLYLQFAADGKAAIATVNDLRNWFLIFAFVAIGLEFSARGLREA
TH_0929|M.thermoresistible__bu --------------------------------------------------
MAB_3433c|M.abscessus_ATCC_199 AVSTIVTLGWLTKG----QVNNLVNVSKWAFLLTFAGVGLNTNFREIARS
MAV_2056|M.avium_104 VVSAIATAGWLTKG----QTANLANVSKWAFLLTFAGVGLNTDIRQIART
MSMEG_0439|M.smegmatis_MC2_155 GWRPIAVFASATVVNIVVALGLATVLFGSFTLD---
TH_0929|M.thermoresistible__bu ------------------------------------
MAB_3433c|M.abscessus_ATCC_199 GWPPLVVAVIGLVVVAAVSLGLVLLTSRVFGWGAHV
MAV_2056|M.avium_104 GWRPLVVAVIGLTVVATVSLGIVLLTSRVFGWGVTT