For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STTTEFAELHNLIGDMRRCVTTLAAKYGDSPAMRRVTNDAERILNDIDRLDIDAEELEMHHGITRTKPGT EKIPVPDTQYGTEFWQDVADEGLGGYR*
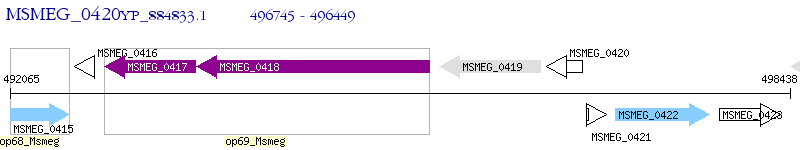
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0420 | - | - | 100% (98) | hypothetical protein MSMEG_0420 |
| M. smegmatis MC2 155 | MSMEG_0163 | - | 2e-16 | 44.94% (89) | hypothetical protein MSMEG_0163 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0256c | - | 7e-28 | 55.10% (98) | hypothetical protein Mb0256c |
| M. gilvum PYR-GCK | Mflv_0392 | - | 2e-36 | 69.47% (95) | hypothetical protein Mflv_0392 |
| M. tuberculosis H37Rv | Rv0250c | - | 7e-28 | 55.10% (98) | hypothetical protein Rv0250c |
| M. leprae Br4923 | MLBr_02557 | - | 5e-27 | 55.67% (97) | hypothetical protein MLBr_02557 |
| M. abscessus ATCC 19977 | MAB_4420 | - | 8e-23 | 50.00% (98) | hypothetical protein MAB_4420 |
| M. marinum M | MMAR_0513 | - | 4e-26 | 54.64% (97) | hypothetical protein MMAR_0513 |
| M. avium 104 | MAV_4907 | - | 6e-29 | 58.51% (94) | hypothetical protein MAV_4907 |
| M. thermoresistible (build 8) | TH_4545 | - | 1e-18 | 63.93% (61) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0288 | - | 9e-27 | 68.42% (76) | hypothetical protein Mvan_0288 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02557|M.leprae_Br4923 ------MYATTELAELHDLIGRMRRSVASFKARYGDSPAMRRIAIDADRI
MAV_4907|M.avium_104 MTSTPGTTGTTEFAELHDLIGGLRRCVSSLRARYGDSPAMRRIVIDADRI
MMAR_0513|M.marinum_M ------MSPTNELAELHDLIGDLRRCVTSLKSRYGDDSGMRRVVIDADRI
Mb0256c|M.bovis_AF2122/97 ------MSTTAELAELHDLVGGLRRCVTALKARFGDNPATRRIVIDADRI
Rv0250c|M.tuberculosis_H37Rv ------MSTTAELAELHDLVGGLRRCVTALKARFGDNPATRRIVIDADRI
MAB_4420|M.abscessus_ATCC_1997 ------MSTKTELTELNQALGSLRRCVHSLQSRYGDLPAVR-------RI
MSMEG_0420|M.smegmatis_MC2_155 ------MSTTTEFAELHNLIGDMRRCVTTLAAKYGDSPAMRRVTNDAERI
TH_4545|M.thermoresistible__bu ---------------------------------------MKRIVNSAERI
Mflv_0392|M.gilvum_PYR-GCK ----MSSTSTTELAQLHDLIGSLRRCVTSLASRYGDSPATRRIVNDAERI
Mvan_0288|M.vanbaalenii_PYR-1 --------------------------MTSLASRYGDSPATRRIVNDAERL
: *:
MLBr_02557|M.leprae_Br4923 LSDIELLDADISELDLARATVQQS--NEKIAIPDTQYDSDFWRDVDDEGV
MAV_4907|M.avium_104 LSDVDLLDTDVSELDLARATVQQS--GEKVVIPDTQYDSEFWRDVDDEGV
MMAR_0513|M.marinum_M LTDIQLLDTDIAELDLDRASVQQS--VEKIAIPDTDYDRDFWRDVDDEGV
Mb0256c|M.bovis_AF2122/97 LTDIELLDTDVSELDLERAAVPQP--SEKIAIPDTEYDREFWRDVDDEGV
Rv0250c|M.tuberculosis_H37Rv LTDIELLDTDVSELDLERAAVPQP--SEKIAIPDTEYDREFWRDVDDEGV
MAB_4420|M.abscessus_ATCC_1997 VNDVERLDIDAADLDLAPHEHRLAG-NEKIQIPDTEYDREFWGDIDDEGV
MSMEG_0420|M.smegmatis_MC2_155 LNDIDRLDIDAEELEMHHGITRTKPGTEKIPVPDTQYGTEFWQDVADEGL
TH_4545|M.thermoresistible__bu QNDIDRLHIDAEEFEMSRGLHRHVPAAEKIPVPDTEYDTNFWHGIDDEGL
Mflv_0392|M.gilvum_PYR-GCK LNDIDRLDIDAEELELARGVSRHHHVAERIPIPDTQYDTEFWRGVDDEGL
Mvan_0288|M.vanbaalenii_PYR-1 LNDIDRLDIDAEELELVRGVSRHPHVAERIAIPDTQYDTEFWRGVDDEGL
.*:: *. * :::: *:: :***:*. :** .: ***:
MLBr_02557|M.leprae_Br4923 GGHSRS
MAV_4907|M.avium_104 GGHHRS
MMAR_0513|M.marinum_M GGHSRY
Mb0256c|M.bovis_AF2122/97 GGHRY-
Rv0250c|M.tuberculosis_H37Rv GGHRY-
MAB_4420|M.abscessus_ATCC_1997 GGYR--
MSMEG_0420|M.smegmatis_MC2_155 GGYR--
TH_4545|M.thermoresistible__bu R-----
Mflv_0392|M.gilvum_PYR-GCK GGTH--
Mvan_0288|M.vanbaalenii_PYR-1 GGTH--