For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHDAQADAARGRAGSARGKYWWLRWVLIAVAVAVLGVELTLVWDQLAKAWKSLVSANVWWVLAAVVAAMA SIHSFAQIQRTLLLSAGVQVRQWRSEAAYYAGNALSTTMPGGPVLSTTFIYRQQRIWGASPLVASWQLVM SGVLQILGLALLGLGGAFMLGASKNPLSLIFSLGGLLALVLLGQAVASRPELIEGIGVKVLHWVNSVRGK PSDTGLAKWREILKQLEAVKLGRRDLTVSFSWSMFNWIADVACLAFACYAAGGHPSLAGVTVAYAAARAV GSIPLMPGGLLVVEAVLVPGLVTSGMTLASAISAMLIYRLISWIFISAIGWVIFFFMFRTETDLDVDRVD QHEPAPERQEPN*
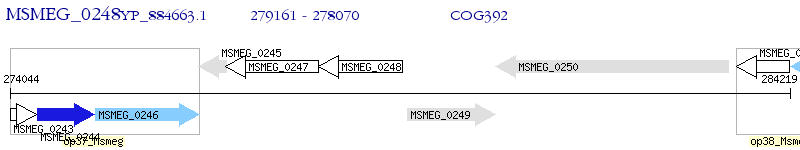
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0248 | - | - | 100% (363) | hypothetical protein MSMEG_0248 |
| M. smegmatis MC2 155 | MSMEG_1199 | - | 2e-08 | 25.00% (256) | hypothetical protein MSMEG_1199 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0210c | - | 1e-147 | 71.07% (363) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_0458 | - | 1e-163 | 78.47% (367) | hypothetical protein Mflv_0458 |
| M. tuberculosis H37Rv | Rv0204c | - | 1e-147 | 70.80% (363) | transmembrane protein |
| M. leprae Br4923 | MLBr_02618 | - | 1e-144 | 75.15% (330) | putative integral membrane protein |
| M. abscessus ATCC 19977 | MAB_4524 | - | 1e-147 | 72.46% (345) | hypothetical protein MAB_4524 |
| M. marinum M | MMAR_0444 | - | 1e-146 | 71.11% (360) | transmembrane protein |
| M. avium 104 | MAV_4970 | - | 1e-149 | 71.35% (363) | hypothetical protein MAV_4970 |
| M. thermoresistible (build 8) | TH_1421 | - | 1e-161 | 75.40% (374) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1094 | - | 1e-146 | 71.11% (360) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0194 | - | 1e-160 | 80.63% (351) | hypothetical protein Mvan_0194 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0248|M.smegmatis_MC2_155 ---------------MSHDAQADAARGRAG---SAR------------GK
TH_1421|M.thermoresistible__bu ---------------VSHDAPATEAPSQLGPQRRAR------------GR
Mflv_0458|M.gilvum_PYR-GCK ---------------MSHDVPASDTLPGSAGAARGR------------GK
Mvan_0194|M.vanbaalenii_PYR-1 ---------------MSHDAPASDAPAGPAGPDRSR------------GK
Mb0210c|M.bovis_AF2122/97 MYRDYGATCTVTLKHVSHDAPARNLRQRVGALPRTRVGAPPAEGVPPRGK
Rv0204c|M.tuberculosis_H37Rv ---------------MSHDAPARNLRQRVGALPRTRVGAPPAEGVPPRGK
MMAR_0444|M.marinum_M ---------------MSFDAPVRKLRQRVGALPRARG------TEAPRGK
MUL_1094|M.ulcerans_Agy99 ---------------MSFDAPVRKLRQRVGALPRARG------TEAPRGK
MLBr_02618|M.leprae_Br4923 ------------------MSPAGGG-----EAPP--------------GK
MAV_4970|M.avium_104 ---------------MPHDAPAHNL-----DLPREQT---------SRGR
MAB_4524|M.abscessus_ATCC_1997 --MPPSDDRDPDRLEGGCTPPDPNGPDSCPDDPEADTAPQP--VIKRRGK
*:
MSMEG_0248|M.smegmatis_MC2_155 YWWLRWVLIAVAVAVLGVELTLVWDQLAKAWKSLVSANVWWVLAAVVAAM
TH_1421|M.thermoresistible__bu YWWLRWAVIAVAVTVLTVEVVLVWDQLAKAWHSLTAANYWWVFAAALAAL
Mflv_0458|M.gilvum_PYR-GCK YWWLRWVVIAVAVIVLAVEIALVRDQLAKAWKSLLSANWVWLIAAVVAAM
Mvan_0194|M.vanbaalenii_PYR-1 YWWLRWALIALAVIVLTVELALVRDQLAKAWKSLVSADWLWVLAAAAAAM
Mb0210c|M.bovis_AF2122/97 YWWLRWAVLAIVAIVLAIEVALGWDQLAKAWVSLYRAKWWWLLAAVAAAG
Rv0204c|M.tuberculosis_H37Rv YWWLRWAVLAIVAIVLAIEVALGWDQLAKAWVSLYRAKWWWLLAAVAAAG
MMAR_0444|M.marinum_M YWWLRWAVLGIVAIVLAVEVALGWDQLAKAWMSMYQAKWWWLLAAMVAAA
MUL_1094|M.ulcerans_Agy99 YWWLRWAVLGIVAIVLAVEVALGWDQLAKAWMSMYQAKWWWLLAAMVAAA
MLBr_02618|M.leprae_Br4923 YWWVRWVVLSLVAIVLVVELALGWDQLAKAWTSVYEANWWWLLAAVLAAA
MAV_4970|M.avium_104 YWWVRWVILGVVAIVLAVEVSLGWDQLAKAWMSMYEANWWWLLASVVAAA
MAB_4524|M.abscessus_ATCC_1997 YWWIRWAILIVLAIVLAVEIGFVSDQLDRAWRSLKTANPWWVLAAALAAM
***:**.:: : . ** :*: : *** :** *: *. *::*: **
MSMEG_0248|M.smegmatis_MC2_155 ASIHSFAQIQRTLLLSAGVQVRQWRSEAAYYAGNALSTTMPGGPVLSTTF
TH_1421|M.thermoresistible__bu ASIHSFAQIQRTLLRSAGVKIRQWRSEAAYYAGNALSTTMPGGPVIATTF
Mflv_0458|M.gilvum_PYR-GCK ASMHSFAQIQRTLLRSAGVRVHQWRSEAAFYAGNALSTTLPGGPVLSATF
Mvan_0194|M.vanbaalenii_PYR-1 ASMHSFAQIQRTLLRSAGVRVHQWRSEAAFYAGNALSTTLPGGPVLSATF
Mb0210c|M.bovis_AF2122/97 ASMHSFAQIQRTLLKSAGVHVKQWRSEAAFYAANSLSTTLPGGPVLSATF
Rv0204c|M.tuberculosis_H37Rv ASMHSFAQIQRTLLKSAGVHVKQWRSEAAFYAANSLSTTLPGGPVLSATF
MMAR_0444|M.marinum_M ASMHSFAQIQRTLLKSAGVHVRQLRSEAAFYAANSLSTTLPGGPVLSATF
MUL_1094|M.ulcerans_Agy99 ASMHSFAQIQRTLLKSAGVHVRQLRSEAAFYAANSLSTTLPGGPVLSATF
MLBr_02618|M.leprae_Br4923 AAVHSFAQIQRTLLKSAGVNVKQLRSEAAFYAANSLSTTLPGGPVLSATF
MAV_4970|M.avium_104 ASMHSFAQIQRTLLKSAGVHVKQLRSEAAFYAANSLSTTLPGGPVLSATF
MAB_4524|M.abscessus_ATCC_1997 ASMHSFAQIQRTLLRSAGVKIRQWRSEAAFYAGNALSTTLPGGPVLSATF
*::*********** ****.::* *****:**.*:****:*****:::**
MSMEG_0248|M.smegmatis_MC2_155 IYRQQRIWGASPLVASWQLVMSGVLQILGLALLGLGGAFMLGASKNPLSL
TH_1421|M.thermoresistible__bu LYRQQRIWGATPLIASWQLVMSGVLQGVGLALLGLGGAVLLGASKNPLSL
Mflv_0458|M.gilvum_PYR-GCK LYRQQRLWGATPLVASWQLVMSGVLQVIGLALLGLGGAFLLGASKNPLSL
Mvan_0194|M.vanbaalenii_PYR-1 IFRQQRLWGASPLVASWQLVMSGALQVIGLALLGLGGAFLLGARENPLSL
Mb0210c|M.bovis_AF2122/97 LLRQQRIWGASTVVASWQLVMSGVLQAVGLALLGLGGAFFLGAKNNPFSL
Rv0204c|M.tuberculosis_H37Rv LLRQQRIWGASTVVASWQLVMSGVLQAVGLALLGLGGAFFLGAKNNPFSL
MMAR_0444|M.marinum_M LLRQQRIWGASTVVASWQLVMSGVLQAVGLALLGLGGAFFLGAKNNPFSL
MUL_1094|M.ulcerans_Agy99 LLRQQRIWGASTVVASWQLVMSGVLQAVGLALLGLGGAFFLGAKNNPFSL
MLBr_02618|M.leprae_Br4923 LLRQQRIWGASTVVASWQLVMSGVLQAVGLALLALGGAFFLGAKNNPFSL
MAV_4970|M.avium_104 LFRQQRLWGASTVVASWQLVMSGVLQAVGLALLGLGGAFLLGAKNNPFSL
MAB_4524|M.abscessus_ATCC_1997 IYRQQRLWGATPVVASWQLVMSGVLQAVGLAILGLGGAFLLGAKTNPFSL
: ****:***:.::*********.** :***:*.****.:*** **:**
MSMEG_0248|M.smegmatis_MC2_155 IFSLGGLLALVLLGQAVASRPELIEGIGVKVLHWVNSVRGKPSDTGLAKW
TH_1421|M.thermoresistible__bu IFTLGGFLALLVLAQAVASRPELIDGIGVRLLTRFNALRGKPATTGLDKW
Mflv_0458|M.gilvum_PYR-GCK IFSVGGFVALILLAQAVATRPELIDGIGARVLTWFNSVRGKDADTGLAKW
Mvan_0194|M.vanbaalenii_PYR-1 IFSLGGFVALILLAQAVATRPELLDGIGARVLRWINSVRGKPTRAGLAKW
Mb0210c|M.bovis_AF2122/97 LFTLGGFVTLLLLAQAVASRPELIEGIGRRVLSWANSVRGRPADAGLPKW
Rv0204c|M.tuberculosis_H37Rv LFTLGGFVTLLLLAQAVASRPELIEGIGRRVLSWANSVRGRPADAGLPKW
MMAR_0444|M.marinum_M LFTLGGFIALLLLAQAVASRPELLEGIGRRVLAWVNSIRGRPAATGMEKW
MUL_1094|M.ulcerans_Agy99 LFTLGGFIALLLLAQAVASRPELLEGIGRRVLAWVNSIRGRPAATGMEKW
MLBr_02618|M.leprae_Br4923 LFALAGFVTLLLLAQAVASRPELIEGIGRRVLSWVNSVRGKSADTGLDKW
MAV_4970|M.avium_104 LFTLGGFVALLLLAQAVASRPELIEGIGSRVLAWVNSVRGRPAETGLDKW
MAB_4524|M.abscessus_ATCC_1997 VFSVGGLLAILLLAQAVASRPEMLDGIGVRVLRWVNDLRGKPPMMGVARW
:*::.*::::::*.****:***:::*** ::* * :**: . *: :*
MSMEG_0248|M.smegmatis_MC2_155 REILKQLEAVKLGRRDLTVSFSWSMFNWIADVACLAFACYAAGGHPSLAG
TH_1421|M.thermoresistible__bu RELLSQLKSVNLGRRDLSNAFSWSLFNWVADVACLAFACYAAGGSPSLAG
Mflv_0458|M.gilvum_PYR-GCK REILSQLEAVQLGRRDLGAAFGWSLFNWVADVACLLFACYATGWHPSIAG
Mvan_0194|M.vanbaalenii_PYR-1 REILTQLESVQLSRRDLGEAFSWSLFNWIADVACLLFACFATGGHPSLAG
Mb0210c|M.bovis_AF2122/97 RETLMQLESVSLGRRDLGVAFGWSLFNWIADVACLGFAAYAAGDHASVGG
Rv0204c|M.tuberculosis_H37Rv RETLMQLESVSLGRRDLGVAFGWSLFNWIADVACLGFAAYAAGDHASVGG
MMAR_0444|M.marinum_M RQILMQLESVSLSRRDLGVAFSWSLFNWIADVACLGFAAYAAGDHASVAG
MUL_1094|M.ulcerans_Agy99 RQILMQLESVSLSRRDLGVAFSWSLFNWIADVACLGFAAYAAGDHASVAG
MLBr_02618|M.leprae_Br4923 RQTLTQLESVSLGRRDLAMAFSWSLFNWIADVACLGFAAYAAGDHASVAG
MAV_4970|M.avium_104 RETLMQLESVSLGRRDLSVAFGWSMFNWIADVACLGFAAYAAGDHASVAG
MAB_4524|M.abscessus_ATCC_1997 REILKQMEAVTLTRRALGEAFGWSLFNWIADVACLAFAAYAVGSRPSLVG
*: * *:::* * ** * :*.**:***:****** **.:*.* .*: *
MSMEG_0248|M.smegmatis_MC2_155 VTVAYAAARAVGSIPLMPGGLLVVEAVLVPGLVTSGMTLASAISAMLIYR
TH_1421|M.thermoresistible__bu VTVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMTLAGAISAMLVYR
Mflv_0458|M.gilvum_PYR-GCK VTVAYAAARAVGSIPLMPGGLLVVEAVLVPGLVSSGMSLAAAISAMLIYR
Mvan_0194|M.vanbaalenii_PYR-1 VTVAYAAARAVGSIPLMPGGLLVVEAVLVPGLVSSGMSLASAISAMLIYR
Mb0210c|M.bovis_AF2122/97 LAVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMPLPSAISAMLIYR
Rv0204c|M.tuberculosis_H37Rv LAVAYAAARAVGTIPLMPGGVLVVEAVLVPGLVSSGMPLPSAISAMLIYR
MMAR_0444|M.marinum_M LTVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMALPNAISAMLIYR
MUL_1094|M.ulcerans_Agy99 LTVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMALPNAISAMLIYR
MLBr_02618|M.leprae_Br4923 LTVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMSLPSAISAMLIYR
MAV_4970|M.avium_104 LTVAYAAARAVGTIPLMPGGLLVVEAVLVPGLVSSGMSLPSAISAMLIYR
MAB_4524|M.abscessus_ATCC_1997 VMVAYAAARAAGSVPLMPGGLLVVEAFLVPGLVSSGMTLPDAISAMLIYR
: ********.*::******:*****.******:***.*. ******:**
MSMEG_0248|M.smegmatis_MC2_155 LISWIFISAIGWVIFFFMFRTETDLDVDR---------------------
TH_1421|M.thermoresistible__bu LISWVFLAAIGWVVFFFMFRTEKDIDPDAGMESERV--------------
Mflv_0458|M.gilvum_PYR-GCK LISWIFISAIGWVVFFFMFRTEQQIDPD---AP-----------------
Mvan_0194|M.vanbaalenii_PYR-1 LISWIFISAIGWVVFFFMFRTAHDVEPDPAAAPTATP-----AEDVAPQL
Mb0210c|M.bovis_AF2122/97 LISWLLIAAIGWVVFFFMFRTESTADSDNDRDPPTDPNLRLVIQPQGTPC
Rv0204c|M.tuberculosis_H37Rv LISWLLIAAIGWVVFFFMFRTESTADSDNDRDPPTDPNLRLVIQPQGTPC
MMAR_0444|M.marinum_M LISWLFIAAIGWVVFFFMFRTEKAADSDAD-DPPTDPNLAIVIPIPGELY
MUL_1094|M.ulcerans_Agy99 LISWLFIAAIGWVVFFFMFRTEKAADSDAD-DPPTDPNLAIVIPIPGELY
MLBr_02618|M.leprae_Br4923 LISWLLIAAIGWVVFFFMFRTENVTDPDD-----------------LGCN
MAV_4970|M.avium_104 LISWLLIAAVGWVVFFFVFRTENIADSDDE--PITGP---LPVLPTPGGP
MAB_4524|M.abscessus_ATCC_1997 AVSWVFISAIGWVIFFFLFRDESQIDPDATPAAAKDP-------------
:**::::*:***:***:** : *
MSMEG_0248|M.smegmatis_MC2_155 -----VDQHEPAPE-RQEPN-------
TH_1421|M.thermoresistible__bu -----APVFSPDPEVRHEPDS------
Mflv_0458|M.gilvum_PYR-GCK ---------PPGNGAEPAPRTDD----
Mvan_0194|M.vanbaalenii_PYR-1 FSPDPDAFHPPDQKSGAPPEDPDR---
Mb0210c|M.bovis_AF2122/97 DDPVETTPQGPAPTPDLRPEGGETPPR
Rv0204c|M.tuberculosis_H37Rv DDPVETTPQGPAPTPDLRPEGGETPPR
MMAR_0444|M.marinum_M EDPAETALQGPLPPPDRGCEGNPGRVG
MUL_1094|M.ulcerans_Agy99 EDPAETALQGPLPPPDRGCEGNPGRVG
MLBr_02618|M.leprae_Br4923 PD--------------QPLD-------
MAV_4970|M.avium_104 PDPTDTALQGPLPPDRSPADPNPDKDV
MAB_4524|M.abscessus_ATCC_1997 ---DATPGAPPANPPKNSSDPPK----