For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
APDVRDMRDARRKDMSPIQPPSNRSPGQRNQGPRPAAVRPRDPQAPARGAARGPQRQFDDRPVEYWPTAA IRAALETDDLTVWQRIVAAIKRDPYGRTARQVEEVLETARPYGVSKALSEVLVRTRDHLEASERAEVAKQ IQAMLRRSKLAPPEFASRVGVSNEEFATYLEGSVSPRASLLLRMQRLSDRFAKIAAQRSGK*
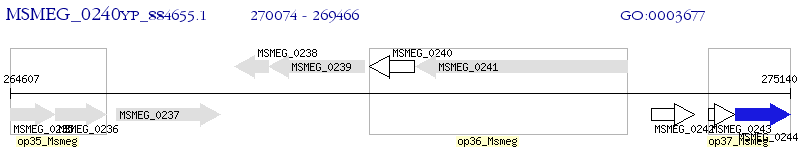
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0240 | - | - | 100% (202) | hypothetical protein MSMEG_0240 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0207c | - | 1e-42 | 59.29% (140) | hypothetical protein Mb0207c |
| M. gilvum PYR-GCK | Mflv_0465 | - | 7e-57 | 71.33% (150) | hypothetical protein Mflv_0465 |
| M. tuberculosis H37Rv | Rv0201c | - | 1e-42 | 59.29% (140) | hypothetical protein Rv0201c |
| M. leprae Br4923 | MLBr_02616 | - | 1e-46 | 55.97% (159) | hypothetical protein MLBr_02616 |
| M. abscessus ATCC 19977 | MAB_4530 | - | 1e-36 | 48.77% (162) | hypothetical protein MAB_4530 |
| M. marinum M | MMAR_0441 | - | 6e-49 | 59.28% (167) | hypothetical protein MMAR_0441 |
| M. avium 104 | MAV_4974 | - | 9e-51 | 52.91% (189) | hypothetical protein MAV_4974 |
| M. thermoresistible (build 8) | TH_2115 | - | 1e-54 | 69.54% (151) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1091 | - | 1e-48 | 58.68% (167) | hypothetical protein MUL_1091 |
| M. vanbaalenii PYR-1 | Mvan_0187 | - | 2e-56 | 70.78% (154) | hypothetical protein Mvan_0187 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0465|M.gilvum_PYR-GCK ------------------MTPGVSQTPQTRR----------------DTG
Mvan_0187|M.vanbaalenii_PYR-1 -----------------MVAPEMSG--KVR-------------------G
MSMEG_0240|M.smegmatis_MC2_155 ------------------MAPDVRDMRDARRKDMSP----IQPPSNRSPG
TH_2115|M.thermoresistible__bu MTAELHRAADRYGPRRESRAPRQPRNGQLRAPQGRAGGADPTRGPNPTGG
MLBr_02616|M.leprae_Br4923 ----------------MTLADDH--RRPAAPP------------------
MAV_4974|M.avium_104 ----------------MTLAPD---RRPASPP------------------
Mb0207c|M.bovis_AF2122/97 ----------------MTLAAEP----HPAPP------------------
Rv0201c|M.tuberculosis_H37Rv ----------------MTLAAEP----HPAPP------------------
MMAR_0441|M.marinum_M ----------------MTLATDQRKPAQVQPP------------------
MUL_1091|M.ulcerans_Agy99 ----------------MTLATDQRKPAQVQPP------------------
MAB_4530|M.abscessus_ATCC_1997 ----------------MTVTSDAPAVSPEVPDEPSG-----------SGA
:
Mflv_0465|M.gilvum_PYR-GCK AENATPPPA-----------------------------------------
Mvan_0187|M.vanbaalenii_PYR-1 EDRRVTPPA-----------------------------------------
MSMEG_0240|M.smegmatis_MC2_155 QRNQGPRPAAVRPRDPQAPARGAARGPQ----------------------
TH_2115|M.thermoresistible__bu PNNPTGGPGEGRPGQGAGQTRAVPGGHEGRAVPGGESRVGQGGHDRSGQS
MLBr_02616|M.leprae_Br4923 -EQPAADQGR----------------------------------------
MAV_4974|M.avium_104 -HRPAPGDQRNRGGGPSAPL------------------------------
Mb0207c|M.bovis_AF2122/97 -QQPTVAWSE----------------------------------------
Rv0201c|M.tuberculosis_H37Rv -QQPTVAWSE----------------------------------------
MMAR_0441|M.marinum_M -HRPDGDNRE----------------------------------------
MUL_1091|M.ulcerans_Agy99 -HRPDGDNRE----------------------------------------
MAB_4530|M.abscessus_ATCC_1997 SGSTSAPPGR----------------------------------------
Mflv_0465|M.gilvum_PYR-GCK --------DTRPVEFWPTAAIRAALESDDLSVWQRIVVAIKRDPYGRTAR
Mvan_0187|M.vanbaalenii_PYR-1 --------DDRPVEFWPTAAIRAALENDDLAVWQRIVVAIKRDPFGRTAR
MSMEG_0240|M.smegmatis_MC2_155 -----RQFDDRPVEYWPTAAIRAALETDDLTVWQRIVAAIKRDPYGRTAR
TH_2115|M.thermoresistible__bu GEARRRPHDNRPVEFWPTSEIRAALENDDLPVWRQIAAAIKRDPYGRTAR
MLBr_02616|M.leprae_Br4923 ------YDPDQPVEFWSTAAIRSALHAGSIEIWKLITAAVKHDPYGRTAH
MAV_4974|M.avium_104 ------PDLDQPVEFWPTAAIRSALQSGDIATWKRIAGALKRDPYGRTAR
Mb0207c|M.bovis_AF2122/97 ------PDVDRRVEFWPTVAIRSALESGDIATWQRIAAALKRDPYGRTAR
Rv0201c|M.tuberculosis_H37Rv ------PDVDRRVEFWPTVAIRSALESGDIATWQRIAAALKRDPYGRTAR
MMAR_0441|M.marinum_M ------HFVDQPVEFWPTAAIRSALQSGDIATWKRIAAALKRDPYGRTAR
MUL_1091|M.ulcerans_Agy99 ------HFVDQPVEFWPTATIRSALQSGDIATWKRIAAALKRDPYGRTAR
MAB_4530|M.abscessus_ATCC_1997 --------GDKPVTHWTTAEILGALQSGDLSDWQAIVHALKRDPFGRTAR
: * .*.* * .**. ..: *: *. *:*:**:****:
Mflv_0465|M.gilvum_PYR-GCK QVEEVLETARPYGVSKAMAEVLTRTREHLEANECAEVARHVRLLLERSGL
Mvan_0187|M.vanbaalenii_PYR-1 QVEEVLETARPYGVSRAMSEVLTRTREHLEANECAEVARHVHLLLERSGL
MSMEG_0240|M.smegmatis_MC2_155 QVEEVLETARPYGVSKALSEVLVRTRDHLEASERAEVAKQIQAMLRRSKL
TH_2115|M.thermoresistible__bu QVEEVIETAQPYGVSKAINEVLARAREALEQSERAEVAKHVRLLLDRSGL
MLBr_02616|M.leprae_Br4923 QVEEVLEGTRPYGICKALGEVLQRARTHLEINERAEVARHVRLLIDRSGL
MAV_4974|M.avium_104 QVEEVLDGTRPYGIAKALWEVLERARAHLEANERAEVARHVRLLIDRSGL
Mb0207c|M.bovis_AF2122/97 QVEEVLEGIPATGIANAFWEVLDRARTHLDANERAEVARQVGLLLDRSGL
Rv0201c|M.tuberculosis_H37Rv QVEEVLEGIPATGIANAFWEVLDRARTHLDANERAEVARQVGLLLDRSGL
MMAR_0441|M.marinum_M QVEEVLGGARPIGISKALWEVLDRARAHLEANERAEVARHVKLLMDRSGL
MUL_1091|M.ulcerans_Agy99 QVEEVLGGARPIGISKALWEVLDRARAHLEANERAEVARHVKLLMDRSGL
MAB_4530|M.abscessus_ATCC_1997 QVEEVVSADELYGISTALQEVLIRARDDLAAAERREAANEINLLFEASGL
*****: *:. *: *** *:* * * *.*..: :: * *
Mflv_0465|M.gilvum_PYR-GCK GEQEFASRIGVPADDFATYLQGSVSPAASLMIRMGRLSDRFARMRAQRPP
Mvan_0187|M.vanbaalenii_PYR-1 GEHEFASRIGVPADDFAAYLRGSVSPPASLMIRMGRLSDRFAKMRAQRPR
MSMEG_0240|M.smegmatis_MC2_155 APPEFASRVGVSNEEFATYLEGSVSPRASLLLRMQRLSDRFAKIAAQRSG
TH_2115|M.thermoresistible__bu AAREFASRIGATGEEFAAYLDGSVSPPASLMIRMRRLSDRFAKMREQRSK
MLBr_02616|M.leprae_Br4923 GHQEFASRIGVAPEGLASYLDGSTSPSAALMVRMRRVSDRFVKVKAARSA
MAV_4974|M.avium_104 GQQEFASRIGVSAEELGSYLDGSTSPTAALMIRIRRLSDRFVKVKSARSA
Mb0207c|M.bovis_AF2122/97 QRQEFASRIGVTAQDLTAYLDGIVSPSASLMIRMRRLSDRFVRAKSVRAA
Rv0201c|M.tuberculosis_H37Rv QRQEFASRIGVTAQDLTAYLDGIVSPSASLMIRMRRLSDRFVRAKSVRAA
MMAR_0441|M.marinum_M GQQEFASRIGVSPEDLAAYLDGSVSPSASLMIRMRRLSDRFVRVKNTRAA
MUL_1091|M.ulcerans_Agy99 GQQEFASRIGVSPEDLAAYLDGSVSPSASLMIRMRRLSDRFVRVKNTRAA
MAB_4530|M.abscessus_ATCC_1997 REAEFASRIGVSGSELSAYLAGSTIPSVAMLVRMRRVARRFGRTRPRG--
*****:*.. . : :** * . * .::::*: *:: ** :
Mflv_0465|M.gilvum_PYR-GCK S---------
Mvan_0187|M.vanbaalenii_PYR-1 D---------
MSMEG_0240|M.smegmatis_MC2_155 K---------
TH_2115|M.thermoresistible__bu RSG-------
MLBr_02616|M.leprae_Br4923 NSD-------
MAV_4974|M.avium_104 ESN-------
Mb0207c|M.bovis_AF2122/97 DS--------
Rv0201c|M.tuberculosis_H37Rv DS--------
MMAR_0441|M.marinum_M DSDQDAASGS
MUL_1091|M.ulcerans_Agy99 DSDQDAASGS
MAB_4530|M.abscessus_ATCC_1997 ----------