For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTARDTQRARVYAAEEFVRTMFDRAAQHSSRAIDFFGTQLTLPPEAKFGSVAAVQRYVDDVIARVGAAPV AVRARRGTKAAHYEIENAVIAVPEQQTTWALRELVVLHELAHHLCATGPDPDAPAHGPQFVATYCELCET VMGPEVGLVLRMVYAKEGVS
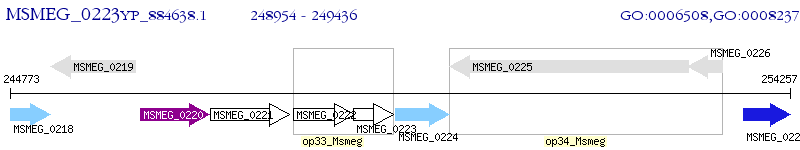
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0223 | - | - | 100% (160) | hypothetical protein MSMEG_0223 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0191 | - | 5e-51 | 63.25% (166) | hypothetical protein Mb0191 |
| M. gilvum PYR-GCK | Mflv_0515 | - | 5e-51 | 63.64% (165) | hypothetical protein Mflv_0515 |
| M. tuberculosis H37Rv | Rv0185 | - | 5e-51 | 63.25% (166) | hypothetical protein Rv0185 |
| M. leprae Br4923 | MLBr_02605 | - | 3e-53 | 62.28% (167) | hypothetical protein MLBr_02605 |
| M. abscessus ATCC 19977 | MAB_4549c | - | 2e-43 | 62.42% (149) | hypothetical protein MAB_4549c |
| M. marinum M | MMAR_0429 | - | 5e-47 | 57.65% (170) | hypothetical protein MMAR_0429 |
| M. avium 104 | MAV_4995 | - | 9e-53 | 64.46% (166) | hypothetical protein MAV_4995 |
| M. thermoresistible (build 8) | TH_0984 | - | 7e-59 | 67.86% (168) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1079 | - | 5e-47 | 57.65% (170) | hypothetical protein MUL_1079 |
| M. vanbaalenii PYR-1 | Mvan_0153 | - | 5e-52 | 64.24% (165) | hypothetical protein Mvan_0153 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0515|M.gilvum_PYR-GCK ------MTRDAQRAKVYAAEGFVRTMFDRAAERGDPVADFFGTHLTLPPE
Mvan_0153|M.vanbaalenii_PYR-1 ------MTRDGQRAKVYAAEGFVRTMFDRAAERGNPVVDFFGTQLTLPPE
TH_0984|M.thermoresistible__bu -VVTEVTVRDTQRAKVYAAEQFVRTMFDRAAEHGNRAVDFFGTRLTLPPE
MSMEG_0223|M.smegmatis_MC2_155 -----MTARDTQRARVYAAEEFVRTMFDRAAQHSSRAIDFFGTQLTLPPE
Mb0191|M.bovis_AF2122/97 MIGADVP-RDSQRARVYAAEAFVRTLFDRVTAHGSPTVEFFGTQLTLPPE
Rv0185|M.tuberculosis_H37Rv MIGADVP-RDSQRARVYAAEAFVRTLFDRVTAHGSPTVEFFGTQLTLPPE
MMAR_0429|M.marinum_M MTAAESSKRDFQRAKVYAAEEFVRTLFDRAAEHGTPSVEFFGTQLTLPPE
MUL_1079|M.ulcerans_Agy99 MTAAESSKRDFQRARVYAAEEFVRTLFDRATEHGTPSVEFFGTQLTLPPE
MAV_4995|M.avium_104 MSPGDSPPRDSQRSRVYAAEEFVRTLFDRAAEHGSPAVEFFGTRLTLPPE
MLBr_02605|M.leprae_Br4923 MNLLDSV-RDAQRSKVYAAEEFIRTLFDRAVEHGSPAVEFFGAQLSLPPE
MAB_4549c|M.abscessus_ATCC_199 ----------------------MRTIFERAAGHSRRDIDFFGARLTLPPE
:**:*:*.. :. :***::*:****
Mflv_0515|M.gilvum_PYR-GCK ARFASAESVQTYVDDVLAHPAVRERWPAAGAVRVRARRGASAAHYESAGG
Mvan_0153|M.vanbaalenii_PYR-1 GRFGSAEAVQSYVDDVLSHPAVQDRWPAAAPLRVRPRRGAAAAHYERADG
TH_0984|M.thermoresistible__bu ARFGSVESVQRYVDEVLDHPAVREQWPSAGPLAVRPRRGVTAAHYERRGT
MSMEG_0223|M.smegmatis_MC2_155 AKFGSVAAVQRYVDDVIAR-------VGAAPVAVRARRGTKAAHYEIEN-
Mb0191|M.bovis_AF2122/97 GRFGSVASVQRYVDDVLALPAVGQNWPTVSPVRVRARRAATAAHYENHGG
Rv0185|M.tuberculosis_H37Rv GRFGSVASVQRYVDDVLALPAVGQNWPTVSPVRVRARRAATAAHYENHGG
MMAR_0429|M.marinum_M GRFASVSSVQSYVDSVLGLPAVRQHWPAVSPLRVRARQAATAAHYENRGG
MUL_1079|M.ulcerans_Agy99 GRFASVSSVQSYVDSVLGLPAVRQHWPAVSPLRVRARQAATAAHYENRGG
MAV_4995|M.avium_104 GRFGSVPSVQRYVDQVLALPAVRQRWPDVPPLRVRPRRAATAAHYENRYG
MLBr_02605|M.leprae_Br4923 ARFGSVAAVQRYVDDVLALQAVRQRWPRMLPLTVRARRAATAAHYENLDG
MAB_4549c|M.abscessus_ATCC_199 ARFASVASVQRYVDDVLAL--VHGRWPAG-PVTVRARRGATAAHYERDGD
.:*.*. :** ***.*: .: **.*:.. *****
Mflv_0515|M.gilvum_PYR-GCK EATIAVP-----HGRDTWALRELVVLHELAHHLCPS-----DPPHGPEFV
Mvan_0153|M.vanbaalenii_PYR-1 AATIAVP-----QGRAVWALRELVVLHEIAHHLCDV-----DPPHGPAFV
TH_0984|M.thermoresistible__bu SAVIAVP-----ERRDTWALRELVVLHEIAHHLCDA-----EPAHGPEFV
MSMEG_0223|M.smegmatis_MC2_155 -AVIAVP-----EQQTTWALRELVVLHELAHHLCATGPDPDAPAHGPQFV
Mb0191|M.bovis_AF2122/97 TGTIAVP----DRHTAGWAMRELVVLHEVAHHLCQV-----PPPHGPEFV
Rv0185|M.tuberculosis_H37Rv TGTIAVP----DRHTAGWAMRELVVLHEVAHHLCQV-----PPPHGPEFV
MMAR_0429|M.marinum_M VGTIAVPGAGPGRGAGDWAMRELVVLHEVAHHLCQV-----PPPHGPEFV
MUL_1079|M.ulcerans_Agy99 VGTIAVPGAGPGRGAGDWAMRELVVLHEVAHHLCQV-----PPPHGPEFV
MAV_4995|M.avium_104 AGVIAVP----DRDTAEWALRELVVLHEVAHHLCRA-----GPPHGPEFV
MLBr_02605|M.leprae_Br4923 AGVIAVP-----GNNADWAMRELVVLHEVAHHLCKD-----PPPHGPEFV
MAB_4549c|M.abscessus_ATCC_199 RAAIAVPD---DRSGSAWAMRELVILHELAHHLCPQ----DGPAHGHDFV
..**** **:****:***:***** *.** **
Mflv_0515|M.gilvum_PYR-GCK AAHCELAGAVMGPEVAHVLRVVYAKEGVR
Mvan_0153|M.vanbaalenii_PYR-1 ATFCELAATVMGPEVAHVLRVVYAREGVK
TH_0984|M.thermoresistible__bu ATFCALAGAVMGPEVGHVLRVVYAKEGVR
MSMEG_0223|M.smegmatis_MC2_155 ATYCELCETVMGPEVGLVLRMVYAKEGVS
Mb0191|M.bovis_AF2122/97 ATVCTLTELVMGPEVGHVFRVVYAQEGVR
Rv0185|M.tuberculosis_H37Rv ATVCTLTELVMGPEVGHVFRVVYAQEGVR
MMAR_0429|M.marinum_M ATMCTLAELVMGPELGHVLRVVYAKEGVR
MUL_1079|M.ulcerans_Agy99 ATMCTLAELVMGPELGHVLRVVYAKEGVR
MAV_4995|M.avium_104 ATICELTELVMGPEVGHVLRVVYAKEGVR
MLBr_02605|M.leprae_Br4923 ATICALTELVMGPELGYVFRVVYAKEGVR
MAB_4549c|M.abscessus_ATCC_199 VLYPELAGLAMGPEVEFVLRTVYAREGAR
. * .****: *:* ***:**.