For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTMLIHHEIFAAHRTAAGHPERPDRYRAVEMALAQSRFDDLLRVEAELAELDVTRYVHSNAYVDALEAA RPTDGFVYLDGGDTMMEPSTWEVVLRGVGGTLQAVDSVLDGTAQNAFVACRPPGHHAETERAMGFCLFNN ISIGARHAQLKHGLSRVAIVDFDVHHGNGTQEIFYSDGSVVYASTHQMPLFPGTGATDETGVGNIFNAPL RAGDGGDELREAFRDRIIPAVDRFEPELIIVSAGFDAHERDPLGSLSMTADDFGWATRELMAAAERHCEG RLVSVLEGGYDLQGLRESVSAHVDELVRG
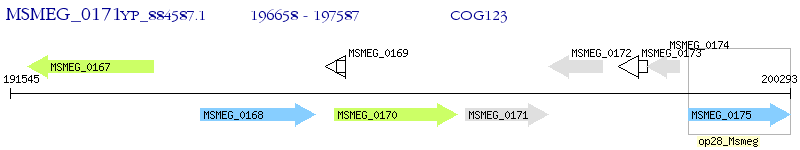
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0171 | - | - | 100% (309) | histone deacetylase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3333 | - | 1e-143 | 78.96% (309) | histone deacetylase superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0171|M.smegmatis_MC2_155 MTTMLIHHEIFAAHRTAAGHPERPDRYRAVEMALAQSRFDDLLRVEAELA
MAV_3333|M.avium_104 MATLLLHHPDFAAHRTAPGHPERPDRYRAVAAALSRPGFDALVRETAEPA
*:*:*:** *******.************ **::. ** *:* ** *
MSMEG_0171|M.smegmatis_MC2_155 ELDVTRYVHSNAYVDALEAARPTDGFVYLDGGDTMMEPSTWEVVLRGVGG
MAV_3333|M.avium_104 ELATTRYVHSNRYVDALEAARPQHGYVYLDGGDTMMEPSTWETALRGVGA
** .******* ********** .*:****************..*****.
MSMEG_0171|M.smegmatis_MC2_155 TLQAVDSVLDGTAQNAFVACRPPGHHAETERAMGFCLFNNISIGARHAQL
MAV_3333|M.avium_104 TLQAVDRVLAGDVQNAFVACRPPGHHAETERAMGFCLFNNISIGARHAQR
****** ** * .************************************
MSMEG_0171|M.smegmatis_MC2_155 KHGLSRVAIVDFDVHHGNGTQEIFYSDGSVVYASTHQMPLFPGTGATDET
MAV_3333|M.avium_104 KHGLMRVAIVDFDVHHGNGTQQIFYSDPSVLYASTHQMPLFPGTGAAAET
**** ****************:***** **:***************: **
MSMEG_0171|M.smegmatis_MC2_155 GVGNIFNAPLRAGDGGDELREAFRDRIIPAVDRFEPELIIVSAGFDAHER
MAV_3333|M.avium_104 GVGNIFNSPLAPGDGGPELRAAFTDRIVPALQAFSPELIIVSAGFDAHER
*******:** .**** *** ** ***:**:: *.***************
MSMEG_0171|M.smegmatis_MC2_155 DPLGSLSMTADDFGWATRELMAAAERHCEGRLVSVLEGGYDLQGLRESVS
MAV_3333|M.avium_104 DPLGSLTMTTDDFGWVTRELMKSAEKLCDGRLVAVLEGGYDLQALADSVT
******:**:*****.***** :**: *:****:*********.* :**:
MSMEG_0171|M.smegmatis_MC2_155 AHVDELVRG
MAV_3333|M.avium_104 AHVGELLKG
***.**::*