For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHILGVDTAITLVAAGAIFLLALALGVWKYRQVMVSDGHRAHVYVDIAHRAALLYAFATLLVAVFVELSV WPAWLNLTAAMVMVFFFVAAIATYIFHGARRDTENQFDPAVPGTGVFMVSLMLGEIGGFAVVFVGFIVGQ LR*
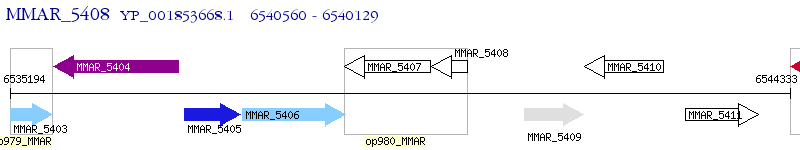
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5408 | - | - | 100% (143) | hypothetical protein MMAR_5408 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3887c | - | 4e-11 | 54.90% (51) | hypothetical protein Mb3887c |
| M. gilvum PYR-GCK | Mflv_0389 | - | 6e-47 | 64.54% (141) | hypothetical protein Mflv_0389 |
| M. tuberculosis H37Rv | Rv3857c | - | 4e-11 | 54.90% (51) | hypothetical protein Rv3857c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4688 | - | 6e-49 | 65.47% (139) | hypothetical protein MAB_4688 |
| M. avium 104 | MAV_0170 | - | 5e-51 | 68.12% (138) | hypothetical protein MAV_0170 |
| M. smegmatis MC2 155 | MSMEG_6442 | - | 1e-37 | 62.50% (112) | integral membrane protein |
| M. thermoresistible (build 8) | TH_2107 | - | 3e-48 | 67.15% (137) | integral membrane protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0295 | - | 2e-45 | 62.77% (137) | hypothetical protein Mvan_0295 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0389|M.gilvum_PYR-GCK ----------MSTPLAVDTALTVLAAGLIFLWALVLGIWKYRQMATSADH
Mvan_0295|M.vanbaalenii_PYR-1 MWIPPVPCCAMGTPLAIDTALTVLAAGLIFIWALILGVWKYRQMATSPDH
TH_2107|M.thermoresistible__bu ----------MRADLGTDTVITLLAAGLIFLWALILGVWKYRQMATAQNH
MAB_4688|M.abscessus_ATCC_1997 -------MIASGYVLSTDTAVTLLAAGLIFLLSLVLGVWKYRQMVTGPNH
MSMEG_6442|M.smegmatis_MC2_155 --------------------MRLLNS-------------KYRQMATSENH
MMAR_5408|M.marinum_M ----------MSHILGVDTAITLVAAGAIFLLALALGVWKYRQVMVSDGH
MAV_0170|M.avium_104 -------MTFGCGSLGLDTKITLLAAGLIFLLALILGVWKYREMMTSENH
Mb3887c|M.bovis_AF2122/97 --------------------------------------------------
Rv3857c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0389|M.gilvum_PYR-GCK LAHPYVDIAHRAALLYSFATMLLAVFVELSAWPTAVNLTAAAVVVAFFVL
Mvan_0295|M.vanbaalenii_PYR-1 LAHPYVDIAHRAALLYAFATMLLAVFVELSAWPTWVNLTSAAVVVAFFVL
TH_2107|M.thermoresistible__bu LAHPYVDIAHRAALLYSFATLLIAVFVELSGWPAAVNLTAAMVVVFFFVV
MAB_4688|M.abscessus_ATCC_1997 LAHPYVDTAHRAALLYSFATLLTAVFVELSAWPSWVNLTAAMMLVFFFVA
MSMEG_6442|M.smegmatis_MC2_155 LAHPYVDMTHRAALLYSFATLLVAAFVELSVWATWVNMTAAMVLAVFFVI
MMAR_5408|M.marinum_M RAHVYVDIAHRAALLYAFATLLVAVFVELSVWPAWLNLTAAMVMVFFFVA
MAV_0170|M.avium_104 LAHPYVDIAHRAALLYSFATLLIAVFVQLSAWPVWVNLAAAGVLVFFFLA
Mb3887c|M.bovis_AF2122/97 ---------MNCALGFDTKPILLA--------------------------
Rv3857c|M.tuberculosis_H37Rv ---------MNCALGFDTKPILLA--------------------------
..** : .:* *
Mflv_0389|M.gilvum_PYR-GCK AIATYIAHGIRRDTTNQFEHVDTTIRIAMAALIVGEIGGFAVLLAGFVAG
Mvan_0295|M.vanbaalenii_PYR-1 AIATYVMHGMRRDTTNQFERIDTTVRVAMAALIVGEIGGTVVLVAGFAAA
TH_2107|M.thermoresistible__bu AIGSYVLHGARRDTTNQFEHSTTALHGAMVALIAGEIGGFAVLLAGFVSA
MAB_4688|M.abscessus_ATCC_1997 AIASYIAHGALRDTTNQFEKPTIGLYLAMALLILGEIGGFSVLLAGFARA
MSMEG_6442|M.smegmatis_MC2_155 AVFAYILHGARRDTTNQFENATPALHAGMYALIVAEIGGFCVLFTGFVAG
MMAR_5408|M.marinum_M AIATYIFHGARRDTENQFDPAVPGTGVFMVSLMLGEIGGFAVVFVGFIVG
MAV_0170|M.avium_104 AILAYIGHGARRDTVNQFERPTAGLRWSMAALIVGEIGGFGVLFAGFVAG
Mb3887c|M.bovis_AF2122/97 ---SYVTHGARRATANQFERPAKGAGVLMALLILGEMAGFAVVVTGVVFG
Rv3857c|M.tuberculosis_H37Rv ---SYVTHGARRATANQFERPAKGAGVLMALLILGEMAGFAVVVTGVVFG
:*: ** * * ***: * *: .*:.* *:..*. .
Mflv_0389|M.gilvum_PYR-GCK QFF-
Mvan_0295|M.vanbaalenii_PYR-1 QFS-
TH_2107|M.thermoresistible__bu QFL-
MAB_4688|M.abscessus_ATCC_1997 QWFS
MSMEG_6442|M.smegmatis_MC2_155 QFF-
MMAR_5408|M.marinum_M QLR-
MAV_0170|M.avium_104 QLW-
Mb3887c|M.bovis_AF2122/97 QLV-
Rv3857c|M.tuberculosis_H37Rv QLV-
*