For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GRFRYIARDDRWEWSDELARMHGYEPGRVRPTADLLVAHKHPDDKAVVAELVDQVCRYGKPCSNRFRIID TGGVVHIVVVLANPLYDGHGSLAGTAGFYVDITEQYEVDMQKRLTQAVTAVNARRAVINQAIGMLMHEYR LDAESAFQVLAKWSQKSNTKLRLLAERVVGDPDGRNVLLNEAADTVGELLRAARNGHG*
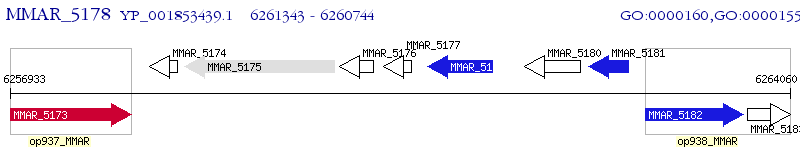
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5178 | - | - | 100% (199) | putative response regulatory protein |
| M. marinum M | MMAR_0736 | - | 8e-44 | 48.84% (172) | putative regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3447 | - | 6e-47 | 49.42% (172) | putative PAS/PAC sensor protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01796 | - | 5e-13 | 32.77% (119) | hypothetical protein MLBr_01796 |
| M. abscessus ATCC 19977 | MAB_2523c | - | 1e-40 | 47.09% (172) | putative transcription antitermination regulator |
| M. avium 104 | MAV_3103 | - | 2e-42 | 51.79% (168) | ANTAR domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_0542 | - | 6e-36 | 42.44% (172) | ANTAR domain-containing protein |
| M. thermoresistible (build 8) | TH_1795 | - | 1e-39 | 46.29% (175) | antar domain protein |
| M. ulcerans Agy99 | MUL_3601 | - | 3e-43 | 48.26% (172) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5727 | - | 2e-41 | 48.26% (172) | putative PAS/PAC sensor protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5178|M.marinum_M --------------------------------------------------
MAV_3103|M.avium_104 -----------------------------------MTVNSNDETVPCAE-
MUL_3601|M.ulcerans_Agy99 MSSPCHLPTPALSGQVPTVDGLQPSSQRSWCALPRALIGSASTLAQCGAG
Mflv_3447|M.gilvum_PYR-GCK --------------------------------------------------
TH_1795|M.thermoresistible__bu --------------------------------------------------
Mvan_5727|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2523c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0542|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01796|M.leprae_Br4923 --------------------------------------------------
MMAR_5178|M.marinum_M --------------------------------------------------
MAV_3103|M.avium_104 ----------PLGSDVVEGDGDLSAVERVLG---------------LGEP
MUL_3601|M.ulcerans_Agy99 VQVRLGSWWLPGGSGAMISDVQQTAHTGEERPDRGTPSFATPISGAVDNH
Mflv_3447|M.gilvum_PYR-GCK --------------------------------------------------
TH_1795|M.thermoresistible__bu ------------------VPGDGSVEDALAG----------------GSP
Mvan_5727|M.vanbaalenii_PYR-1 ------------------MPSDDSVEQALAG----------------GAP
MAB_2523c|M.abscessus_ATCC_199 -----------MSDELSEEEADSELAQALAG----------------GSP
MSMEG_0542|M.smegmatis_MC2_155 ------------MTTAPEDDDTAAAVQALTG----------------GTP
MLBr_01796|M.leprae_Br4923 --------------------------------------------------
MMAR_5178|M.marinum_M --MGRFRYIARDDRWEWSDELARMHGYEPGRVRPTADLLVAHKHPDDKAV
MAV_3103|M.avium_104 QRVGRFRFFLDGHRWEWSDAVARMHGYRPGQVQPSTELLLQHKHPDDRER
MUL_3601|M.ulcerans_Agy99 LSIGSFRFWFIGQRWEWSDEVAGMHGYEPGAVVPTTKLLLSHKHPDDRAH
Mflv_3447|M.gilvum_PYR-GCK MSVGSFNYLRSDDRWVWSDAVAAMHGYAPGTVEPTTELVLSHKHPDDKAT
TH_1795|M.thermoresistible__bu QRVGWFRFYFADERWEWSPQVQQMHGYEPGTVTPTTELVLSHKHPDDYAQ
Mvan_5727|M.vanbaalenii_PYR-1 QRVGWFRFYFADERWEWSEQVQILHGYEPGTVTPTTDLVLSHKHPDDRQQ
MAB_2523c|M.abscessus_ATCC_199 QHVGWFRFYFADERWEWSPQVAQMHGYGPDEITPTTEIILSHKHPDDYRQ
MSMEG_0542|M.smegmatis_MC2_155 QQMGWFRFYFADQHWEWSAEVQRMHGYEPGTVTPTTELVLSHKHPDDRDQ
MLBr_01796|M.leprae_Br4923 ------------------------------MSSLTTDLMLTHRHLNDRGQ
::.::: *:* :*
MMAR_5178|M.marinum_M VAELVDQVCRYGKPCSNRFRIIDTGGVVHIVVVLANPLYDGHGSLAGTAG
MAV_3103|M.avium_104 VAAVLDQVMR-GKPFSSRHRIVDTAGRTHWVVVAGDRMLDDSGALAGTSG
MUL_3601|M.ulcerans_Agy99 VQDLLDYALRSGESFSSRHRFVDTAGVVHDAIVVADRMVNESGVVQGTSG
Mflv_3447|M.gilvum_PYR-GCK VAQLIDRARREGAVFSGRHRIIDTHGKTHVVIVVGDTLTDDAGEVVGTTG
TH_1795|M.thermoresistible__bu MVATLEQIRRSHNTFSTRHRIVDKHGDVHSVVVVGDQLCDENGAVIGTHG
Mvan_5727|M.vanbaalenii_PYR-1 VAATLDEMRRTHRTFSTRHRIVDTRGDTHDVVVIGDQLLDDAGRLIGTHG
MAB_2523c|M.abscessus_ATCC_199 VAATLQEIRRTAQAFSTRHRIIDVQQHVHHVVVIGDQLCDDNGVVVGTHG
MSMEG_0542|M.smegmatis_MC2_155 VAVDINDMMTGRRAFGRRHRIIDTSGTLRQVIVVSDQLHGFDGDVIGIRG
MLBr_01796|M.leprae_Br4923 VAATIDEILNTHKLFSTRHRIIDTSENVDNVIVIADQLFDDRGATIGTYD
: :: . *.*::* .:* .: : . * * .
MMAR_5178|M.marinum_M FYVDITEQYEVDMQKRLTQAVTAVNARRAVINQAIGMLMHEYRLDAESAF
MAV_3103|M.avium_104 FYVDVTDS----LHSDITNVLSAVADSRARIEQAKGVLMAAYGISAERAF
MUL_3601|M.ulcerans_Agy99 YYIDLTNTFDETRQEVLDEALPDLFENRAAIEQAKGVLMAVYRVSAEEAF
Mflv_3447|M.gilvum_PYR-GCK FYVDITDGFHADVQRSVSRHVTNIDENRATIQQAVGMIRMAYGVSADRAF
TH_1795|M.thermoresistible__bu FYVDVTPNIAVADD-LVSAAVAQIAENRATIEQAKGMLMLIYRIGADAAF
Mvan_5727|M.vanbaalenii_PYR-1 FYVDITPTDTQAKA-MVTAALAEITENRAVIEQVKGMLMLIYRVDDEAAF
MAB_2523c|M.abscessus_ATCC_199 FYVDVTSSIRQERENSLTVAVVEIAENRSVIEQAKGMLMLVYRLNADAAF
MSMEG_0542|M.smegmatis_MC2_155 FYIDVTPPRSRQEDAVITAEVAKITERRAVIEQVKGMLMLIYGIDENAAF
MLBr_01796|M.leprae_Br4923 FYIDVSALPEQVHEGIVIARLAKSTQNRAGIEQTKSMLMLIDGITYDTPF
:*:*:: : : *: *:*. .:: : : .*
MMAR_5178|M.marinum_M QVLAKWSQKSNTKLRLLAERVVGDPDGRN--VLLNEAADTVGELLRAARN
MAV_3103|M.avium_104 DILVWRSQETNLKLRDVAGRFL-DAVASK--ASPGMQRQVDHALLTLE--
MUL_3601|M.ulcerans_Agy99 RVLQWRSQETNTKLRALAKQVVAEVASLPP-VSALVQSQFDHLLLTVHER
Mflv_3447|M.gilvum_PYR-GCK DVLKWRSQETNAKLRHIADRLVAELSAAP--LSPESRARVDHILLTAHLD
TH_1795|M.thermoresistible__bu DLLKWRSQETNVKLRMLAEQLIEDFTGLDYQDELPTRGTFDHLLLTAHQR
Mvan_5727|M.vanbaalenii_PYR-1 ELLRWRSQETNVKLRLLADQLLADYTTLSYDETLPPRATFDHLLLTAHQR
MAB_2523c|M.abscessus_ATCC_199 DVLKWRSQENNVKLRALAQQVVVDFGTLDFDKTLPARSVYDRLLLTAHQR
MSMEG_0542|M.smegmatis_MC2_155 DLLKWLSQENNVKLRVLADQVAKDFTALG-DTGVLSRSKFDQLLLTAARR
MLBr_01796|M.leprae_Br4923 NLLKYAVPGN--QHQAAAG-------------------------------
:* . : : *
MMAR_5178|M.marinum_M GHG----------
MAV_3103|M.avium_104 -------------
MUL_3601|M.ulcerans_Agy99 IPAESAS------
Mflv_3447|M.gilvum_PYR-GCK G------------
TH_1795|M.thermoresistible__bu TNSDGRRAGIS--
Mvan_5727|M.vanbaalenii_PYR-1 VNRAGDTVDG---
MAB_2523c|M.abscessus_ATCC_199 IPR----------
MSMEG_0542|M.smegmatis_MC2_155 AVSSDDDSMSTSA
MLBr_01796|M.leprae_Br4923 -------------