For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GPTRRRDLAAAAIGAAVAGYPMVLVLFRSFPPLTVWTGLSLLGVAIAEALWARFVRAKINDGEIGDGSGQ LHPLAVARSLMVAKASAWVGALVLGWWIAVLVYLLPRRSWLRVAAEDMTGALVAAASALALIVAALWLQH CCKSPQEPPERGEGAEG*
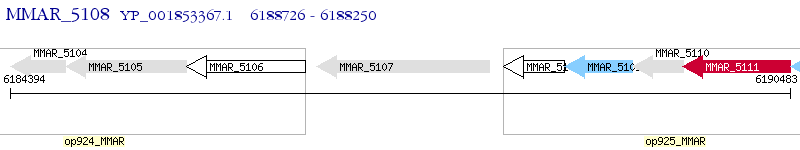
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5108 | - | - | 100% (158) | hypothetical protein MMAR_5108 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3635c | - | 4e-70 | 77.71% (157) | hypothetical protein Mb3635c |
| M. gilvum PYR-GCK | Mflv_1421 | - | 2e-57 | 63.92% (158) | hypothetical protein Mflv_1421 |
| M. tuberculosis H37Rv | Rv3605c | - | 4e-70 | 77.71% (157) | hypothetical protein Rv3605c |
| M. leprae Br4923 | MLBr_00227 | - | 5e-70 | 78.98% (157) | hypothetical protein MLBr_00227 |
| M. abscessus ATCC 19977 | MAB_0538 | - | 6e-45 | 55.33% (150) | hypothetical protein MAB_0538 |
| M. avium 104 | MAV_0548 | - | 6e-69 | 77.71% (157) | hypothetical protein MAV_0548 |
| M. smegmatis MC2 155 | MSMEG_6100 | - | 3e-59 | 67.53% (154) | hypothetical protein MSMEG_6100 |
| M. thermoresistible (build 8) | TH_2443 | - | 2e-53 | 66.03% (156) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4187 | - | 3e-87 | 99.37% (158) | hypothetical protein MUL_4187 |
| M. vanbaalenii PYR-1 | Mvan_5367 | - | 8e-58 | 65.19% (158) | hypothetical protein Mvan_5367 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1421|M.gilvum_PYR-GCK MGLTRKRDLAAAVAAAAIAGYLLMYVVYRVFPPITVWTGMSLLGVAVALA
Mvan_5367|M.vanbaalenii_PYR-1 MGPTRTRDLAAAATVTAIAGYLLVHVAYRWFPPIDVWTGVSLLGVAAAVG
MMAR_5108|M.marinum_M MGPTRRRDLAAAAIGAAVAGYPMVLVLFRSFPPLTVWTGLSLLGVAIAEA
MUL_4187|M.ulcerans_Agy99 MGPTRRRDLAAAAIGAAVAGYPMVLVLFRSFPPLTVWTGLSLLGVAIAEA
Mb3635c|M.bovis_AF2122/97 MGPTRKRDLTAAVVGAAAVGYLLVAVLYRWFPPITVWTGLSLLAVAVAEA
Rv3605c|M.tuberculosis_H37Rv MGPTRKRDLTAAVVGAAAVGYLLVAVLYRWFPPITVWTGLSLLAVAVAEA
MLBr_00227|M.leprae_Br4923 MGPTRKRDLTAAMIGAAVVGYLLVLVLYRWFPPITVWTGLSLLAVAIPEA
MAV_0548|M.avium_104 MGPTRKRDLTAAVVGAAVVGYLLVQGLYRWFPPITVWTGLSLLAVAVIEA
MSMEG_6100|M.smegmatis_MC2_155 MGPTRKRDLVGAAVAAALLSYLLIHALYRWFPPITVWTGLSLLGVAAFEA
TH_2443|M.thermoresistible__bu MGPTRLRYLAAAAVAAAVVGYITVLLLYRWFPPITVLSGISLLILALPEA
MAB_0538|M.abscessus_ATCC_1997 MGTTRFRDLLVAGVVTAIFCYFFVSAAYGSLPPIPLLAGVSLLVLAVAEA
** ** * * * :* * : : :**: : :*:*** :* .
Mflv_1421|M.gilvum_PYR-GCK GWAFLVRARIRDGRIGVGAGRLHPLAVARSVVIAKASAWMGSVVLGWWLA
Mvan_5367|M.vanbaalenii_PYR-1 GWAFYVRARIRDGQIGVGQGRLHPLAVARSVVIAKASAWMGSVVFGWWLA
MMAR_5108|M.marinum_M LWARFVRAKINDGEIGDGSGQLHPLAVARSLMVAKASAWVGALVLGWWIA
MUL_4187|M.ulcerans_Agy99 LWARFVRAKINDGEIGDGSGQLHPLAVARSLMVAKASAWVGALVLGWWIA
Mb3635c|M.bovis_AF2122/97 LWARYVRVKISDGEIGDGPGWLHPLVVARSLMVAKASAWVGALVTGWWIG
Rv3605c|M.tuberculosis_H37Rv LWARYVRVKISDGEIGDGPGWLHPLVVARSLMVAKASAWVGALVTGWWIG
MLBr_00227|M.leprae_Br4923 LWARYVRTKISDGEIGDGPGWLHPLAVAHSLMVAKASAWVGALVLGWWVG
MAV_0548|M.avium_104 LWARYVRTKINDGEIGSGPGWLHPLAVARSLMVAKASAWVGALMLGWWIG
MSMEG_6100|M.smegmatis_MC2_155 GWGFYVRAKINDGQIGVGAGRLDPLTVARSVVIAKASAWVGALVAGWWVG
TH_2443|M.thermoresistible__bu GWAFYVRAKINNGEIGIGGGRLDPLWVARSVYIAKASAWVGALMFGWWAG
MAB_0538|M.abscessus_ATCC_1997 GWAFYVRNRVNDGQIGVGAGRLPALAVARSVVVAKASAWLGTLMTGWWMA
*. ** :: :*.** * * * .* **:*: :******:*::: *** .
Mflv_1421|M.gilvum_PYR-GCK VVIYLLPRRASLRVAAEDTAGAVIAALCALALVIAALWLQYCCTSPDDSA
Mvan_5367|M.vanbaalenii_PYR-1 VVVYLLPRRATLRVAAEDTPGAVVAGLCALALVVAAMWLQHCCRSPEDQS
MMAR_5108|M.marinum_M VLVYLLPRRSWLRVAAEDMTGALVAAASALALIVAALWLQHCCKSPQEPP
MUL_4187|M.ulcerans_Agy99 VLVYLLPRRSWLRVAAEDMTGALVAAASALALIVAALWLQNCCKSPQEPP
Mb3635c|M.bovis_AF2122/97 VLAYFLPRRSWLRAAAEDTTGTVVAAGSALALVVAALWLQHCCKSPQDPT
Rv3605c|M.tuberculosis_H37Rv VLAYFLPRRSWLRAAAEDTTGTVVAAGSALALVVAALWLQHCCKSPQDPT
MLBr_00227|M.leprae_Br4923 VLVYFLPRWPWLRVADKDTSGTVVAALSALALLVAALWLQHCCKSPQDPT
MAV_0548|M.avium_104 VLVYFLPRRSWLRAASEDTSGAVVAAVSALALLVAALWLQHCCKSPPDSG
MSMEG_6100|M.smegmatis_MC2_155 VLVYLLPRRTTIRVAGEDTPGVVVAALCALALAIAGLWLQHCCKSPEEPP
TH_2443|M.thermoresistible__bu TLCYLLPLRDTLRVAAADTPGAVVAALSGLALGAAALWLQHCCKSPPEPP
MAB_0538|M.abscessus_ATCC_1997 MLVYILPRRAHLAAAAADTLGVVIATGCALALVVAGLWLQHCCKSPPDPP
: *:** : .* * *.::* ..*** *.:*** ** ** :
Mflv_1421|M.gilvum_PYR-GCK PEADPAPG
Mvan_5367|M.vanbaalenii_PYR-1 ENADAATG
MMAR_5108|M.marinum_M ERGEGAEG
MUL_4187|M.ulcerans_Agy99 ERGEGAEG
Mb3635c|M.bovis_AF2122/97 EHADGAES
Rv3605c|M.tuberculosis_H37Rv EHADGAES
MLBr_00227|M.leprae_Br4923 EHGEGAEN
MAV_0548|M.avium_104 EHGEGAET
MSMEG_6100|M.smegmatis_MC2_155 EDGDAVPE
TH_2443|M.thermoresistible__bu DGADPAPE
MAB_0538|M.abscessus_ATCC_1997 --ATPAR-
. .