For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KLGLQLGYWGAQPPENAGELVAAAEGAGFDAVFTAEAWGSDAYTPLAWWGSSTQRLRLGTSVIQLSARTP TACAMAALTLDHLSGGRHILGLGVSGPQVVEGWYGQRFPKPLARTREYVDILRQVWAREGVVTSAGPHYP LPLTGPGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIFYTPRMADMYNEWLDEGFA RPGARRSRADFEICATAQVVVTEDRPAAFAAIKPFVALYMGGMGAEGTNFHADVYRRMGYAEVVDDVTKL FRSNRKDEAAEIIPDELVDDAVIVGDADYVRKQIEVWEAAGVTMMVVSARSVEQVRELAGLVGG*
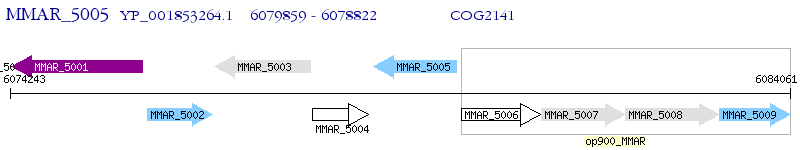
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5005 | - | - | 100% (345) | coenzyme F420-dependent oxidoreductase |
| M. marinum M | MMAR_1111 | - | 6e-68 | 40.52% (348) | coenzyme F420-dependent oxidoreductase |
| M. marinum M | MMAR_0154 | - | 2e-67 | 42.42% (330) | coenzyme F420-dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3550c | - | 0.0 | 90.09% (343) | coenzyme F420-dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1539 | - | 1e-175 | 85.71% (343) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3520c | - | 0.0 | 90.09% (343) | coenzyme F420-dependent oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 1e-178 | 87.72% (342) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4166c | - | 1e-161 | 79.82% (342) | putative luciferase-like protein |
| M. avium 104 | MAV_0639 | - | 0.0 | 89.80% (343) | FMN-dependent monooxygenase |
| M. smegmatis MC2 155 | MSMEG_5920 | - | 1e-175 | 84.55% (343) | FMN-dependent monooxygenase |
| M. thermoresistible (build 8) | TH_2264 | - | 1e-166 | 86.34% (322) | POSSIBLE COENZYME F420-DEPENDENT OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4079 | - | 0.0 | 98.55% (345) | coenzyme F420-dependent oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5219 | - | 1e-170 | 83.67% (343) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5005|M.marinum_M -------MKLGLQLGYWGAQPPENAGELVAAAEGAGFDAVFTAEAWGSDA
MUL_4079|M.ulcerans_Agy99 -------MKLGLQLGYWGAQPPENAGELVAAAEEAGFDAVFTAEAWGSDA
Mb3550c|M.bovis_AF2122/97 ---MEAGMKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
Rv3520c|M.tuberculosis_H37Rv ---MEAGMKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
Mflv_1539|M.gilvum_PYR-GCK -------MKLGLQLGYWGAQPPENHAELVAAAEGAGFDTVFTAEAWGSDA
Mvan_5219|M.vanbaalenii_PYR-1 -------MKLGLQLGYWGAQAPTNHAELVATAEEVGFDTVFTAEAWGSDA
TH_2264|M.thermoresistible__bu ----------------------------VAAAEEAGFDTVFTAEAWGSDA
MAV_0639|M.avium_104 -------MKLGLQLGYWGAGPPENHGELVAAAEEAGYDAVFTAEAWGSDA
MSMEG_5920|M.smegmatis_MC2_155 -------MKLGLQLGYWAAQPPSNHAELVAAAEDAGFDTVFTAEAWGSDA
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWGSDA
MAB_4166c|M.abscessus_ATCC_199 -------MKFGLQLGYWSASPPENAGELVAAAEEGGFDAVFTAEAWGSDA
**:** *:*:***.*******
MMAR_5005|M.marinum_M YTPLAWWGSSTQRLRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
MUL_4079|M.ulcerans_Agy99 YTPLAWWGSSTQRLRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Mb3550c|M.bovis_AF2122/97 YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Rv3520c|M.tuberculosis_H37Rv YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Mflv_1539|M.gilvum_PYR-GCK YTPLAWWGRETTRMRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Mvan_5219|M.vanbaalenii_PYR-1 YTPLAWWGRETTRMRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
TH_2264|M.thermoresistible__bu FTPLAWWGRQTTRMRLGTSVVQLSARTPTACAMASLTLDHLSGGRHILGL
MAV_0639|M.avium_104 YTPLAWWGSSTSRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
MSMEG_5920|M.smegmatis_MC2_155 FTPLAWWGRETERLRLGTSVVQMSARTPTACAMASLTLDHLSGGRHILGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
MAB_4166c|M.abscessus_ATCC_199 YTPLAWWGSDTSRIRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
:******* .* *:******:*:***********:***************
MMAR_5005|M.marinum_M GVSGPQVVEGWYGQRFPKPLARTREYVDILRQVWAREGVVTSAGPHYPLP
MUL_4079|M.ulcerans_Agy99 GVSGPQVVEGWYGQRFPKPLARTREYVDILRQVWAREGVVTSDGPHYPLP
Mb3550c|M.bovis_AF2122/97 GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
Rv3520c|M.tuberculosis_H37Rv GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
Mflv_1539|M.gilvum_PYR-GCK GVSGPQVVEGWYGAKFPKPLARTREYVDILRQVWAREAPVHSDGPHYPLP
Mvan_5219|M.vanbaalenii_PYR-1 GVSGPQVVEGWYGAKFPKPLARTREYIDILRQVWAREAPVHSDGPHYPLP
TH_2264|M.thermoresistible__bu GVSGPQVVEGWYGQRFPKPLARTREYIDIIRQVWAREAPVTSAGPHYPLP
MAV_0639|M.avium_104 GVSGPQVVEGWYGQRFPKPLARTREYIDIIRQVWAREKPVTSDGPHYPLP
MSMEG_5920|M.smegmatis_MC2_155 GVSGPQVVEGWYGQKFPKPLARTREYIDIIRQVWAREAPVTSDGPHYPLP
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFPKPLSRTREYIDIVRQVWAREAPVVSDGQHYPLP
MAB_4166c|M.abscessus_ATCC_199 GVSGPQVVEGWYGQPFPKPLARTREYIDIIRQVLAREAPVRSDGPHYPLP
************* *****:*****:**:*** *** * * * ** **
MMAR_5005|M.marinum_M LTGPGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
MUL_4079|M.ulcerans_Agy99 LTGAGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
Mb3550c|M.bovis_AF2122/97 LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
Rv3520c|M.tuberculosis_H37Rv LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
Mflv_1539|M.gilvum_PYR-GCK LTGEGTTGLGKNLKPITHPLRSDIPVMLGAEGPKNVALAAEICDGWLPIF
Mvan_5219|M.vanbaalenii_PYR-1 LTGEGTTGLGKNLKPITHPLRADIPVMLGAEGPKNVALAAEICDGWLPIF
TH_2264|M.thermoresistible__bu LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEIADGWLPIF
MAV_0639|M.avium_104 LTGTGTTGLGKALKPITHPLRADIPIFLGAEGPKNVALAAEICDGWLPIF
MSMEG_5920|M.smegmatis_MC2_155 LTGEGTTGLGKPLKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
MLBr_00348|M.leprae_Br4923 LTGAGATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
MAB_4166c|M.abscessus_ATCC_199 LTGDKATGLGKPLKPIVHPLRSDIPIFLGAEGPKNVALTAEIADGWLPMF
*** :***** ****.****:***::***********:***.*****:*
MMAR_5005|M.marinum_M YTPRMADMYNEWLDEGFARPGARRSRADFEICATAQVVVTEDRPAAFAAI
MUL_4079|M.ulcerans_Agy99 YTPRMADMYNEWLDEGFARPGARRSRADFEICATAQVVVTEDRPAAFAAI
Mb3550c|M.bovis_AF2122/97 YSPRMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAGI
Rv3520c|M.tuberculosis_H37Rv YSPRMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAGI
Mflv_1539|M.gilvum_PYR-GCK YSPRIAGMYNEWLDEGFARPGARRTRETFEICATAQVVVTDDRPAIMELM
Mvan_5219|M.vanbaalenii_PYR-1 YSPRIAAMYNEWLDEGFARPGARHTRETFEICATAQVVVTDDRAEMMELM
TH_2264|M.thermoresistible__bu YSPRLADMYNEWLDEGFARPTARRSREDFEICATAQVVITDDRAGTMEMI
MAV_0639|M.avium_104 YTPRLADMYNEWLDEGFARPGARRSRADFEICATANIVITEDRGAAFAAM
MSMEG_5920|M.smegmatis_MC2_155 YTPRMADTYNAWLDEGFARPGARRTRETFEICATANIVITEDRAAAFAAM
MLBr_00348|M.leprae_Br4923 FSPRMADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAGI
MAB_4166c|M.abscessus_ATCC_199 YAPRLADMYNEWLDEGFSRPGARRSREDFEIAATAQVIVTEDRRSVLDQL
::**:* ** ******:*. **::* ***.*:*::::*:** : :
MMAR_5005|M.marinum_M KPFVALYMGGMGAEGTNFHADVYRRMGYAEVVDDVTKLFRSNRKDEAAEI
MUL_4079|M.ulcerans_Agy99 KPFVALYMGGMGAEGTNFHADAYRRMGYAEVVDDVTKLFRSNRKDEAAEI
Mb3550c|M.bovis_AF2122/97 KPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAEI
Rv3520c|M.tuberculosis_H37Rv KPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAEI
Mflv_1539|M.gilvum_PYR-GCK KPHLALYMGGMGAEDTNFHADVYRRMGYAEVVDDVTRLFRSDRKDEAATV
Mvan_5219|M.vanbaalenii_PYR-1 KPHLALYMGGMGAEDTNFHADVYRRMGYAEVVDDVTKLFRSGRKDEAAKV
TH_2264|M.thermoresistible__bu KPFLALYMGGMGAEEMNFHADVYRRMGYSEVVDDVTRLFRSGRKDEAAKV
MAV_0639|M.avium_104 KPYIALYMGGMGAEDTNFHADVYRRMGYAEVVDDVTKLFRSGRKDEAAKI
MSMEG_5920|M.smegmatis_MC2_155 KPYLALYMGGMGAEDTNFHADVYRRMGYGDVVDEVTKLFRSGRKDEAAKV
MLBr_00348|M.leprae_Br4923 KPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEI
MAB_4166c|M.abscessus_ATCC_199 KPFSALYIGGMGAVELNFHAEVYRRMGYGEVVDEVTELFRTNRKDKAAEA
**. ***:***** ****:.****** :***:** ***:.:**:**
MMAR_5005|M.marinum_M IPDELVDDAVIVGDADYVRKQIEVWEAAGVTMMVVSARSVEQVRELAGLV
MUL_4079|M.ulcerans_Agy99 IPDELVDDAVIVGDADYVRKQVEVWEAAGVTMMVVSARSVEQVRELAGLV
Mb3550c|M.bovis_AF2122/97 IPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVTAGSAEQVRDLAALV
Rv3520c|M.tuberculosis_H37Rv IPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVTAGSAEQVRDLAALV
Mflv_1539|M.gilvum_PYR-GCK IPDELVDDSAIVGDLAYVKEQITAWEAAGVTMMVVGARSPEQIRDLADLV
Mvan_5219|M.vanbaalenii_PYR-1 IPDELVDDSAIVGDLGYVQEQIKAWEAAGVTMMVVGARSPEQIRDLAALV
TH_2264|M.thermoresistible__bu IPDELVDDAAIVGDIDYVRKQIAAWEAAGVTMMVVGARSVEQIRDLAALI
MAV_0639|M.avium_104 IPDEVIDDAAIVGDVDYVRKQIKAWEAAGVTMMVVSGRSTEQVRELATLI
MSMEG_5920|M.smegmatis_MC2_155 IPDEVVDDAAIVGDVSYVREQIKVWEAAGVTTMVVSGRSPEHIRELAALL
MLBr_00348|M.leprae_Br4923 IPNELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF
MAB_4166c|M.abscessus_ATCC_199 IPDELVLDTTIVGTEAEVREQIKAWEAAGVTMLVVGCRSVEHVKQLSALT
**:*:: .: *** * :*: .*.***** ::* * *::::*: *
MMAR_5005|M.marinum_M GG
MUL_4079|M.ulcerans_Agy99 GG
Mb3550c|M.bovis_AF2122/97 --
Rv3520c|M.tuberculosis_H37Rv --
Mflv_1539|M.gilvum_PYR-GCK --
Mvan_5219|M.vanbaalenii_PYR-1 --
TH_2264|M.thermoresistible__bu --
MAV_0639|M.avium_104 --
MSMEG_5920|M.smegmatis_MC2_155 --
MLBr_00348|M.leprae_Br4923 --
MAB_4166c|M.abscessus_ATCC_199 --