For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSMLDRPVKARTFSGLARRRRLVNSLATVLVTLSMLVALTPLIWVLGSVIAKGFKAVSSSAWWTHSQAGM TAFVAGGGAYHAIVGTVLQGLVCAALSIPIGIMVAIYLVEYDGGGLMGRLVSFTVDILTGVPSIVAALFL YALWVGVLGFPRSEFAVSLALVLLMLPVIVRATEEMLRIVPVDLREAGYALGLPKWKTIVRIVIPTGLSG IITGILLALARVMGETAPLLILVGYARTMNFDIFSGYMGSLPGMMYDQTTMGAGMNPVPTDRLWGAALTL ILVIAIINIGARVLTRILVAK*
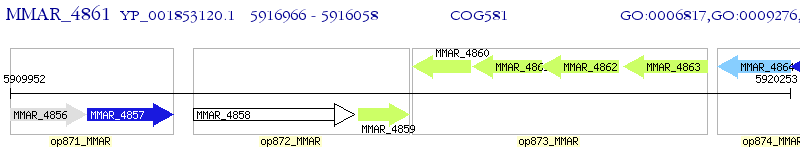
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4861 | pstA | - | 100% (302) | phosphate-transport integral membrane ABC transporter, PstA |
| M. marinum M | MMAR_4578 | pstA1 | 5e-89 | 56.01% (291) | phosphate-transport integral membrane ABC transporter PstA1 |
| M. marinum M | MMAR_4579 | pstC2 | 9e-19 | 27.46% (295) | phosphate-transport integral membrane ABC transporter PstC2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0953 | pstA1 | 8e-91 | 57.00% (293) | phosphate ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1663 | - | 6e-94 | 57.14% (301) | phosphate ABC transporter, inner membrane subunit PstA |
| M. tuberculosis H37Rv | Rv0930 | pstA1 | 8e-91 | 57.00% (293) | phosphate ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 5e-89 | 56.25% (288) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_0748 | - | 1e-105 | 61.59% (302) | phosphate ABC transporter permease |
| M. avium 104 | MAV_0766 | pstA | 1e-136 | 79.14% (302) | phosphate ABC transporter, permease protein PstA |
| M. smegmatis MC2 155 | MSMEG_5780 | pstA | 1e-128 | 74.17% (302) | phosphate ABC transporter, permease protein PstA |
| M. thermoresistible (build 8) | TH_0381 | pstA | 1e-128 | 73.42% (301) | phosphate ABC transporter, permease protein PstA |
| M. ulcerans Agy99 | MUL_0439 | pstA | 1e-164 | 98.68% (302) | phosphate-transport integral membrane ABC transporter, PstA |
| M. vanbaalenii PYR-1 | Mvan_5090 | - | 6e-94 | 57.62% (302) | phosphate ABC transporter, inner membrane subunit PstA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4861|M.marinum_M -----MTSMLDRPVKARTFSGLARRRRLVNSLATVLVTLSMLVALTPLIW
MUL_0439|M.ulcerans_Agy99 -----MTSMLDRPVKARTFSGLARRRRLVNSLATVLVTLSMLVALAPLIR
MAV_0766|M.avium_104 -----MTSMLDRPLKSRTFSPLSRRRRAANSVATVLVSVSLLVAVTPLVM
MSMEG_5780|M.smegmatis_MC2_155 -MTTSMTSTLEQPVKAPAFQGVSLRRRLTDHTATVLVTTSVLIALIPLLW
TH_0381|M.thermoresistible__bu MATPTTTAELDAPVKPTTFQPVSLRRKAVNHMATVLVTASVLIALIPLVW
MAB_0748|M.abscessus_ATCC_1997 -----MTATLDRPVKEPAFHPLSGRRKATNAIATVLVSSAVLIALVPLVW
Mb0953|M.bovis_AF2122/97 MSPSTSIEALDQPVKPVVFRPLTLRRRIKNSVATTFFFTSFVVALIPLVW
Rv0930|M.tuberculosis_H37Rv MSPSMSIEALDQPVKPVVFRPLTLRRRIKNSVATTFFFTSFVVALIPLVW
MLBr_02093|M.leprae_Br4923 ----MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPLVW
Mflv_1663|M.gilvum_PYR-GCK -----MTTTLEKAVKGPTFHPVSASRKFKNGLATTLFAGSFVVAMIPLVW
Mvan_5090|M.vanbaalenii_PYR-1 -----MTATLDEAVKGPTFHPLSAARKFKNGLATTLFTASFVVAMIPLVW
*: .:* .: :: *: : ** :. :.::*: **:
MMAR_4861|M.marinum_M VLGSVIAKGFKAVSSSAWWTHSQAGMTAFVAGGGAYHAIVGTVLQGLVCA
MUL_0439|M.ulcerans_Agy99 VLGSVIAKGFKAVTSSAWWTHSQAGMTAFVAGGGAYHAIVGTVLQGLVCA
MAV_0766|M.avium_104 VLCSVVVKGFRAITSTVWWSHSQAGMTAFVTGGGAYHALVGTVLQGLVCA
MSMEG_5780|M.smegmatis_MC2_155 VLYSVITKGIHAVTSPTWFTNSQAGMTAFSPGGGVYHAIVGTVLQGLVCS
TH_0381|M.thermoresistible__bu VLVSVFVKGIGVVTNTTWWTHSQAGMTAFAAGGGAYHAIVGTLLQGLVCA
MAB_0748|M.abscessus_ATCC_1997 VLYTVFDRGFAAILSADWWFKSQNMMTNRIAGGGAYHAIVGTLFQGLFCA
Mb0953|M.bovis_AF2122/97 LLWVVIARGWFAVTRSGWWTHSLRGVLPEQFAGGVYHALYGTLVQAGVAA
Rv0930|M.tuberculosis_H37Rv LLWVVIARGWFAVTRSGWWTHSLRGVLPEQFAGGVYHALYGTLVQAGVAA
MLBr_02093|M.leprae_Br4923 LLSVVVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVVAA
Mflv_1663|M.gilvum_PYR-GCK LLYTVVAKGISAIGSSTWWMNSLAGVLPEEFAGGVYHAIYGTLIQAAIAA
Mvan_5090|M.vanbaalenii_PYR-1 LIYTVLAKGVAAIVSSTWWTNSLAGVLPEEFAGGVYHAIYGTLIQAAIAA
:: *. :* .: *: :* : .**.***: *::.*. ..:
MMAR_4861|M.marinum_M ALSIPIGIMVAIYLVEYDGGGLMGRLVSFTVDILTGVPSIVAALFLYALW
MUL_0439|M.ulcerans_Agy99 ALSIPIGIMVAIYLVEYDRGGLMGRLVSFTVDILTGVPSIVAALFLYALW
MAV_0766|M.avium_104 AISIPIGLMVAIYLVEYGGGTPLGRLASFMVDILSGVPSIVAALFVYALC
MSMEG_5780|M.smegmatis_MC2_155 LISIPIGVMVAVYLVEYAGGSRLGKITTFMVDILTGVPSIVAALFIYALW
TH_0381|M.thermoresistible__bu AISIPIGVFVGIYLVEYGAGTRLGKLTTFMVDILTGVPSIVAALFIYALW
MAB_0748|M.abscessus_ATCC_1997 IISVPIGIFVAIYLVEYGRNSRLAKLTTFMVDILTGVPSIVAALFIYALW
Mb0953|M.bovis_AF2122/97 VLAVPLGLMTAVYLVEYGTG-RMSRVTTFTVDVLAGVPSIVAALFVFSLW
Rv0930|M.tuberculosis_H37Rv VLAVPLGLMTAVYLVEYGTG-RMSRVTTFTVDVLAGVPSIVAALFVFSLW
MLBr_02093|M.leprae_Br4923 IMAVPLGSMTAVYLVEYGSG-PFVRVTTFMVDVLAGVPSIVAALFIFCLW
Mflv_1663|M.gilvum_PYR-GCK AISVPLGVMAAVFLVEYGGG-RLAKVTTFMVDILAGVPSIVAALFVFALW
Mvan_5090|M.vanbaalenii_PYR-1 LLSVPLGVMAAVYLVEYGQG-RFAKVTTFMVDILAGVPSIVAALFIFALW
:::*:* :..::**** . : ::.:* **:*:**********::.*
MMAR_4861|M.marinum_M VGVLGFPRSEFAVSLALVLLMLPVIVRATEEMLRIVPVDLREAGYALGLP
MUL_0439|M.ulcerans_Agy99 VGVLGFPRSEFAVSLALVLLMLPVIVRATEEMLRIVPVDLREAGYALGLP
MAV_0766|M.avium_104 VATLGLPRSEFAVSLALVLLMLPVIVRATEEMLRIVPVDLREASYALGIT
MSMEG_5780|M.smegmatis_MC2_155 VATLGFQRSGFAVSLSLVLLMIPVIVRATEEMLRIVPMDLREASYALGVP
TH_0381|M.thermoresistible__bu VATLGFGRSGLAVSLSLVLLMVPVIVRSTEEMLRIVPMDLREAAYALGVP
MAB_0748|M.abscessus_ATCC_1997 VATLGLPRSGLAVSLALVLLMIPVVVRSSEEMLKIVPNDLREASYALGVP
Mb0953|M.bovis_AF2122/97 IATLGFQQSAFAVALALVLLMLPVVVRAGEEMLRLVPDELREASYALGVP
Rv0930|M.tuberculosis_H37Rv IATLGFQQSAFAVALALVLLMLPVVVRAGEEMLRLVPDELREASYALGVP
MLBr_02093|M.leprae_Br4923 IATLGFQQSAFAVSLALVLLMLPVVVRSTEEILRLVPDELREACYALGIP
Mflv_1663|M.gilvum_PYR-GCK IATLGFEQSAFAVSLALVLLMLPVVVRNTEEMLKLVPDELREASYALGIP
Mvan_5090|M.vanbaalenii_PYR-1 IATLGFEQSAFAVSLALVLLMLPVVVRNTEEMLKLVPDELREASYALGVP
:..**: :* :**:*:*****:**:** **:*::** :**** ****:.
MMAR_4861|M.marinum_M KWKTIVRIVIPTGLSGIITGILLALARVMGETAPLLILVGYARTMNFDIF
MUL_0439|M.ulcerans_Agy99 KWKTIVRIVIPTGLSGIITGILLALARVMGETAPLLILVGYARTMNFDIF
MAV_0766|M.avium_104 KWKTIARVVLPTGLSGIVTGILLAMARVMGETAPLLILVGYAQSMNFDIF
MSMEG_5780|M.smegmatis_MC2_155 KWKTIVRIVIPTALSGIVTGVMLSLARVMGETAPLLVLVGYAQAMNFDIF
TH_0381|M.thermoresistible__bu KWKTIVRIVIPTALSGIVTGIMLALARVMGETAPLLVLVGYAQAMNFDMF
MAB_0748|M.abscessus_ATCC_1997 KWKTISHIVLPTALSGVVTGVMLALARVMGETAPLLVLVGYSKLINYDMF
Mb0953|M.bovis_AF2122/97 KWKTIVRIVAPIAMPGIVSGILLSIARVVGETAPVLVLVGYSHSINLDVF
Rv0930|M.tuberculosis_H37Rv KWKTIVRIVAPIAMPGIVSGILLSIARVVGETAPVLVLVGYSHSINLDVF
MLBr_02093|M.leprae_Br4923 KWKTIVRIVFPIATPGIISGVLLSIARVIGETAPVLVLVGYSRSINFDIF
Mflv_1663|M.gilvum_PYR-GCK KWKTIVRIVVPTALPGMISGILLALARVMGETAPVLVLVGYSRSINMDAF
Mvan_5090|M.vanbaalenii_PYR-1 KWKTIARIVVPTALPGMISGILLALARVMGETAPVLVLVGYSRSINMDAF
***** ::* * . .*:::*::*::***:*****:*:****:: :* * *
MMAR_4861|M.marinum_M SGYMGSLPGMMYDQTTMGAGMNPVPTDRLWGAALTLILVIAIINIGARVL
MUL_0439|M.ulcerans_Agy99 SGYMGSLPGMMYDQTTMGAGMNPVPTDRLWGAALTLILVIAIINIGARVL
MAV_0766|M.avium_104 SGFMGTLPGMMYNQASAGAGINPIPTDRLWGAALTLILVIATINIVARVI
MSMEG_5780|M.smegmatis_MC2_155 SGFMGSLPGMMYDQISAGAGANPVPTDRLWGAALTLILLIALLNVGARFV
TH_0381|M.thermoresistible__bu TGFMGSLPGMMYDQTSAGAGANPIPTDRLWGAALTLIVLIALLNIGARFL
MAB_0748|M.abscessus_ATCC_1997 GGEMASLPGMMLDQRSG--SVVGLAESRLWGAALTLILLVAVLNVIAKLI
Mb0953|M.bovis_AF2122/97 HGNMASLPLLIYTELTNPEHAGFL---RVWGAALTLIIVVATINLAAAMI
Rv0930|M.tuberculosis_H37Rv HGNMASLPLLIYTELTNPEHAGFL---RVWGAALTLIIVVATINLAAAMI
MLBr_02093|M.leprae_Br4923 DGNMASLPLLIYTELTNPEYAGFL---RVWGAALTLIILVAGINLFGAAF
Mflv_1663|M.gilvum_PYR-GCK EGNMASLPLLIYTELVNPEAAGVL---RVWGAALTLILIIAALYIGAAFI
Mvan_5090|M.vanbaalenii_PYR-1 EGNMASLPLLIYTELVNPEAAGVL---RVWGAALTLILIIAALYIGAAFI
* *.:** :: : : *:********:::* : : . .
MMAR_4861|M.marinum_M TRILVAK-----
MUL_0439|M.ulcerans_Agy99 TRILVAK-----
MAV_0766|M.avium_104 TKFLGARKS---
MSMEG_5780|M.smegmatis_MC2_155 AKIFAPKKV---
TH_0381|M.thermoresistible__bu AKLFAPKKV---
MAB_0748|M.abscessus_ATCC_1997 SRFFAPKKI---
Mb0953|M.bovis_AF2122/97 RFVATRRR----
Rv0930|M.tuberculosis_H37Rv RFVATRRRRLPL
MLBr_02093|M.leprae_Br4923 RFLATRRCSGCR
Mflv_1663|M.gilvum_PYR-GCK NRYLTRNRI---
Mvan_5090|M.vanbaalenii_PYR-1 NRYLMRNRV---
.