For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGEKTTRPRLAQGREHIVATVDEIPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHQGGPLCSGRARGR TMVDESAPGDAVMVRDLEFIYCPWHQWGFELATGTTAVKPEWSIRTYPVRVIGRDVLVMA
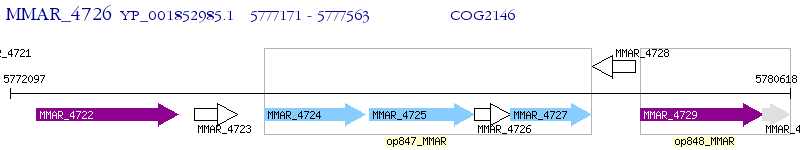
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4726 | - | - | 100% (130) | hypothetical protein MMAR_4726 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2488 | - | 5e-68 | 89.84% (128) | Rieske (2Fe-2S) domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0928 | - | 4e-69 | 89.76% (127) | Rieske (2Fe-2S) domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4809 | - | 2e-69 | 92.74% (124) | Rieske (2Fe-2S) domain-containing protein |
| M. thermoresistible (build 8) | TH_0525 | - | 3e-66 | 85.38% (130) | Rieske |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4166 | - | 2e-68 | 88.28% (128) | Rieske (2Fe-2S) domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4809|M.smegmatis_MC2_155 -----MSDIEK---------------------RP-RLAQGREHVVATVDE
TH_0525|M.thermoresistible__bu -----MTAEQRSAPQQNTPRESTRQSPGQNGSQPKRLAQGREHVVAKVDE
Mflv_2488|M.gilvum_PYR-GCK -----MNNEKT----------------------P-RLAQGREHVVATVDE
Mvan_4166|M.vanbaalenii_PYR-1 -----MDKDKT----------------------P-RLAQGREHVVATVDE
MMAR_4726|M.marinum_M -----MGEKTTR----------------------PRLAQGREHIVATVDE
MAV_0928|M.avium_104 MASETEGAQTKRP--------------------EPRLAQGREHVVATVDE
********:**.***
MSMEG_4809|M.smegmatis_MC2_155 IPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHEGGPLCSGRARGRTVV
TH_0525|M.thermoresistible__bu IPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHEGGPLCSGRPRGRTIV
Mflv_2488|M.gilvum_PYR-GCK IPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHEGGPLCSGRARGRNIV
Mvan_4166|M.vanbaalenii_PYR-1 IPPGTHKLVPIGRHGVGVYNVNGTFHAIANYCPHEGGPLCSGRARGRNIV
MMAR_4726|M.marinum_M IPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHQGGPLCSGRARGRTMV
MAV_0928|M.avium_104 IPPGTHKLVPIGRHGVGVYNVNGTFYAIANYCPHQGGPLCSGRPRGRTIV
*************************:********:********.***.:*
MSMEG_4809|M.smegmatis_MC2_155 DESAPGDAVMVRDMEYIYCPWHQWGFELATGTTAVKPEWSIRTYPVRVVG
TH_0525|M.thermoresistible__bu DDSVPGNAVMVRDMEFIYCPWHQWGFELATGTTAVKPEWSIRTYPVRVVG
Mflv_2488|M.gilvum_PYR-GCK DESVPGDAVMVRDQEYIFCPWHQWGFELATGTTAVKPEWSIRTYPVRVVG
Mvan_4166|M.vanbaalenii_PYR-1 DESVPGDAVMVRDMEFIFCPWHQWGFELATGTTAVKPEWSIRTYPVRVVG
MMAR_4726|M.marinum_M DESAPGDAVMVRDLEFIYCPWHQWGFELATGTTAVKPEWSIRTYPVRVIG
MAV_0928|M.avium_104 DETAPGDSVMVRDLEYIYCPWHQWGFELATGTTAVKPEWSIRTYPVRVVG
*::.**::***** *:*:******************************:*
MSMEG_4809|M.smegmatis_MC2_155 DEVLVTA
TH_0525|M.thermoresistible__bu DEVLVTA
Mflv_2488|M.gilvum_PYR-GCK NDVLVQA
Mvan_4166|M.vanbaalenii_PYR-1 NDVLVQA
MMAR_4726|M.marinum_M RDVLVMA
MAV_0928|M.avium_104 NDVVVQA
:*:* *