For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPFEDSTESSATAVAEWPEELAPTLTGAVALAREAVEEFSGPEAVGDYLGVSYEDPNAATHRFLAHLPG YQGWQWAAVVASYVGADRVTVSEVVLVPGPTALLAPAWVPWEQRVRPGDLSPGDLLAPAKDDPRLVPGYA ASGDAQVDETAAEVGLGRRWVMSGWGRAEAAQRWHDGDCGPDSPMARSTKRVCRDCGFFLPLAGSLGAMF GVCGNELAADGRVVDKQYGCGAHSDTPAPAGTGSPMYEPYDDGVLDVVEKPAESSD*
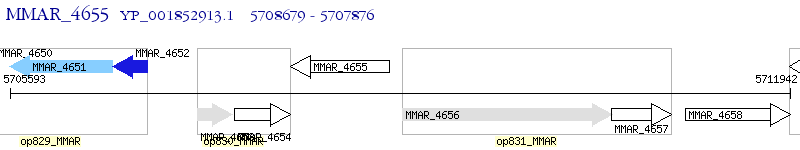
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4655 | - | - | 100% (267) | hypothetical protein MMAR_4655 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0901 | - | 1e-132 | 83.40% (265) | hypothetical protein Mb0901 |
| M. gilvum PYR-GCK | Mflv_1712 | - | 1e-106 | 71.31% (244) | hypothetical protein Mflv_1712 |
| M. tuberculosis H37Rv | Rv0877 | - | 1e-132 | 83.40% (265) | hypothetical protein Rv0877 |
| M. leprae Br4923 | MLBr_02142 | - | 1e-123 | 79.23% (260) | hypothetical protein MLBr_02142 |
| M. abscessus ATCC 19977 | MAB_0876 | - | 1e-86 | 62.96% (243) | hypothetical protein MAB_0876 |
| M. avium 104 | MAV_1007 | - | 1e-125 | 80.30% (264) | hypothetical protein MAV_1007 |
| M. smegmatis MC2 155 | MSMEG_5691 | - | 1e-110 | 69.08% (262) | hypothetical protein MSMEG_5691 |
| M. thermoresistible (build 8) | TH_1376 | - | 1e-105 | 71.13% (239) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0273 | - | 1e-159 | 99.25% (267) | hypothetical protein MUL_0273 |
| M. vanbaalenii PYR-1 | Mvan_5039 | - | 1e-104 | 70.42% (240) | hypothetical protein Mvan_5039 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1712|M.gilvum_PYR-GCK ----------------MTEPDQQADPTGVEPAPTPEPAT-------PSPE
Mvan_5039|M.vanbaalenii_PYR-1 ----------------MTEPDPQATEPGHETGHAPDEAAPEPTFDGPAPD
MSMEG_5691|M.smegmatis_MC2_155 ----------------MTEPADRAEGSDLGSAAAADAPG-----------
TH_1376|M.thermoresistible__bu ------------------XPENPG-------AEVADPRV---------AE
MMAR_4655|M.marinum_M ----------------MTGPFEDSTES--SATAVAEWPE----------E
MUL_0273|M.ulcerans_Agy99 ----------------MTGPFEDSTES--SATAVAEWPE----------E
Mb0901|M.bovis_AF2122/97 ----------------MTGPTEES-----AVATVADWPE----------G
Rv0877|M.tuberculosis_H37Rv ----------------MTGPTEES-----AVATVADWPE----------G
MLBr_02142|M.leprae_Br4923 ----------------MTGPVEDS-----AVATVAEWPE----------E
MAV_1007|M.avium_104 MARQPSIGTAMQTQDSVTTPLEES-----SMATAGEWPE----------G
MAB_0876|M.abscessus_ATCC_1997 -----------------------------MDITISGRD------------
Mflv_1712|M.gilvum_PYR-GCK LEAVLLGAVDDARAAIVEFSGEDSVGEYLGAG--FEDSTSATHRFLAELP
Mvan_5039|M.vanbaalenii_PYR-1 LEALLLGAVEEARAAIVEFSGDGTVGEYLGAG--FEDPSSATHRFLAEMP
MSMEG_5691|M.smegmatis_MC2_155 LESVLRGAVDVARAALTEFSGADTVGEYLGVT--LEDPSSATHRFLANMP
TH_1376|M.thermoresistible__bu LEELLLGAVEQARAAVVEFSGADTVGDYLGAA--LEDSTAATHRFLAKLP
MMAR_4655|M.marinum_M LAPTLTGAVALAREAVEEFSGPEAVGDYLGVS--YEDPNAATHRFLAHLP
MUL_0273|M.ulcerans_Agy99 LAPTLTGAVALAREAVEEFSGAEAVGDYLGVS--YEDPNAATHRFLAHLP
Mb0901|M.bovis_AF2122/97 LAAVLRGAADQARAAVVEFSGPEAVGDYLGVS--YEDGNAATHRFIAHLP
Rv0877|M.tuberculosis_H37Rv LAAVLRGAADQARAAVVEFSGPEAVGDYLGVS--YEDGNAATHRFIAHLP
MLBr_02142|M.leprae_Br4923 LAAVLTNAADDARAAIEEFSGSVTVGDYLGVG--YEDPNAATHRFLAHLP
MAV_1007|M.avium_104 LAAVLTGAADQARAAVVEFSGAETVGDYLGVG--YEDPYAATHRFLAHLP
MAB_0876|M.abscessus_ATCC_1997 ---ALYTCIEQARTAIVEFSG-ETVGKYLGASSEYSDLHALTHRFEAELP
* . ** *: **** :**.***. .* : **** *.:*
Mflv_1712|M.gilvum_PYR-GCK GYRGWQWAVVVAACPGAGRATISEVVLVPGPTALLAPQWVPWEERVRPGD
Mvan_5039|M.vanbaalenii_PYR-1 GYRGWQWAVVVAACPGAAHATISEVVLVPGPTALLAPKWVPWDERIRPGD
MSMEG_5691|M.smegmatis_MC2_155 GYRGWQWAVVVAAYPGADRATISELVLVPGPTALLAPKWVPWQERIRPGD
TH_1376|M.thermoresistible__bu GYRGWQWAVVVAGYPGTDQATISEVVLVPGPTALVAPKWVPWQERIRPGD
MMAR_4655|M.marinum_M GYQGWQWAAVVASYVGADRVTVSEVVLVPGPTALLAPAWVPWEQRVRPGD
MUL_0273|M.ulcerans_Agy99 GYQGWQWAAVVASYVGADRVTVSEVVLVPGPTALLAPAWVPWEQRVRPGD
Mb0901|M.bovis_AF2122/97 GYQGWQWAVVVASYSGADHATISEVVLVPGPTALLAPDWVPWEQRVRPGD
Rv0877|M.tuberculosis_H37Rv GYQGWQWAVVVASYSGADHATISEVVLVPGPTALLAPDWVPWEQRVRPGD
MLBr_02142|M.leprae_Br4923 GYQGWQWAVVVAAYPGAEHATVSEVVLVPGPTALLAPEWVPWEQRVRPGD
MAV_1007|M.avium_104 GYQGWQWAVVVAAYPGADHATISEVVLVPGPTALLAPEWVPWEQRVRPGD
MAB_0876|M.abscessus_ATCC_1997 GYRGWHWEVVMAAAPGATVATVSEVVLVPGAEALRPPNWIPWEERIRPGD
**:**:* .*:*. *: .*:**:*****. ** .* *:**::*:****
Mflv_1712|M.gilvum_PYR-GCK LSPGDLLAPPADDPRLAPGYAATGDPQIDEVAVEVGLGRRQVLSLWGRND
Mvan_5039|M.vanbaalenii_PYR-1 LSPGDLLAPPAEDPRLVPGYTATGDPQIDEVAVEVGLGRRQVLSLWGRND
MSMEG_5691|M.smegmatis_MC2_155 LGPGDLLAPPPDDPRLVPGYSATGDPLIDETALELGFGRRQVLSEWGRAA
TH_1376|M.thermoresistible__bu LGPGDLLAPAVDDPRLVPGHVATGDPAIDEVALELGLGRRWVLSEFGRAE
MMAR_4655|M.marinum_M LSPGDLLAPAKDDPRLVPGYAASGDAQVDETAAEVGLGRRWVMSGWGRAE
MUL_0273|M.ulcerans_Agy99 LSPGDLLAPAKDDPRLVPGYAASGDAQVDETAAEVGLGRRWVMSGWGRAE
Mb0901|M.bovis_AF2122/97 LSPGDLLAPAKDDPRLVPGYTASGDAQVDETAAEIGLGRRWVMSAWGRAQ
Rv0877|M.tuberculosis_H37Rv LSPGDLLAPAKDDPRLVPGYTASGDAQVDETAAEIGLGRRWVMSAWGRAQ
MLBr_02142|M.leprae_Br4923 LGPGDLLAPTSEDLRLVPGYNASGDPAVDEIAAEIGLGRRWVMSVWGRSA
MAV_1007|M.avium_104 LSPGDLLAPAANDPRLVPGYTASGDPQVDETAAEIGLGRRWVMSAEGRAE
MAB_0876|M.abscessus_ATCC_1997 LGPGDLLAPPPNDPRLVPGYTDSGDPQVTETAGEIGLGRKQVLSLAGRID
*.*******. :* **.**: :**. : * * *:*:**: *:* **
Mflv_1712|M.gilvum_PYR-GCK TAQRWHDGEYGPGSAMARATWRMCRDCGYYVPLGGALGVMFGVCANEYAA
Mvan_5039|M.vanbaalenii_PYR-1 TAQRWHDGDYGPGSAMARATRRVCRDCGFYLPLGGALGVLFGVCANEYAA
MSMEG_5691|M.smegmatis_MC2_155 AAQRWHDGEFGPNSPMARSTKRVCRDCGFYLPLGGALGRMFGVCANELSA
TH_1376|M.thermoresistible__bu AARRWHSGEYGPHSAMARATRRVCRDCGFYLPLAGVMGRMFGVCANEFSA
MMAR_4655|M.marinum_M AAQRWHDGDCGPDSPMARSTKRVCRDCGFFLPLAGSLGAMFGVCGNELAA
MUL_0273|M.ulcerans_Agy99 AAQRWHDGDCGPDSPMARSTKRVCRDCGFFLPLAGSLGVMFGVCGNELAA
Mb0901|M.bovis_AF2122/97 SAQRWHDGDYGPGSAMARSTKRVCRDCGFFLPLAGSLGAMFGVCGNELSA
Rv0877|M.tuberculosis_H37Rv SAQRWHDGDYGPGSAMARSTKRVCRDCGFFLPLAGSLGAMFGVCGNELSA
MLBr_02142|M.leprae_Br4923 AAERWHGGDYGPDSPMARSTKRVCRDCGFFLSLVGSLGAMFGVCGNEMSA
MAV_1007|M.avium_104 AAERWHTGAYGPDSPMARSTKRVCRDCGFFLPLSGSLGRMFGVCGNELSA
MAB_0876|M.abscessus_ATCC_1997 AAQRWFDGEWGPDAEMARATRRVCRSCGFYLPLAGSLGVAFGVCANSMSA
:*.**. * ** : ***:* *:**.**:::.* * :* ****.*. :*
Mflv_1712|M.gilvum_PYR-GCK DGHVVDAEFGCGAHSDTPAAAGTGSPLYDPYDDGVLDVVDRPQASGQPTE
Mvan_5039|M.vanbaalenii_PYR-1 DGHVVDAEYGCGAHSDTPAAPGNGSPLFDPYDDGVLDLVEKAD-------
MSMEG_5691|M.smegmatis_MC2_155 DGHVVDSEYGCGAHSDTPAPPGTGSPLYEPFDDGVLDLAD----------
TH_1376|M.thermoresistible__bu DGRVVDAEYGCGAHSDTPRPPGTGSPLYEPYDDGLIEITESVEQT-----
MMAR_4655|M.marinum_M DGRVVDKQYGCGAHSDTPAPAGTGSPMYEPYDDGVLDVVEK------PAE
MUL_0273|M.ulcerans_Agy99 DGRVVDKQYGCGAHSDTPAPAGTGSPMYEPYDDGVLDVVEK------PAE
Mb0901|M.bovis_AF2122/97 DGHVVDRQYGCGAHSDTTAPAGGSTPIYEPYDDGVLDIIEK------PAE
Rv0877|M.tuberculosis_H37Rv DGHVVDRQYGCGAHSDTTAPAGGSTPIYEPYDDGVLDIIEK------PAE
MLBr_02142|M.leprae_Br4923 DGHVVDKLYGCGAHSDTPAPAGSGSSVYEPYDDGVLDILEPSTVQL-PES
MAV_1007|M.avium_104 DGHVVDMHYGCGAHSDTPAPAGTGSPAYEPYDDGLLDVTQTPVEPTPPPE
MAB_0876|M.abscessus_ATCC_1997 DGRVVHIEYGCGAHSDTPQPVNSAMPLYEPFDDGVLDLAD----------
**:**. :********. . . . . ::*:***:::: :
Mflv_1712|M.gilvum_PYR-GCK SPAEVSEESSTESPVEAPAEAPAESPVETPAES
Mvan_5039|M.vanbaalenii_PYR-1 ---------------------------------
MSMEG_5691|M.smegmatis_MC2_155 ---------------------------------
TH_1376|M.thermoresistible__bu ---------------------------------
MMAR_4655|M.marinum_M SSD------------------------------
MUL_0273|M.ulcerans_Agy99 SSD------------------------------
Mb0901|M.bovis_AF2122/97 S--------------------------------
Rv0877|M.tuberculosis_H37Rv S--------------------------------
MLBr_02142|M.leprae_Br4923 PAD------------------------------
MAV_1007|M.avium_104 TPQAEPEPETETPS-------------------
MAB_0876|M.abscessus_ATCC_1997 ---------------------------------