For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDTATSALKPAKALSSPSSLSPQVVIDNDEIFRAHVDGKLSVALKAPLDTQRALSIAYTPGVAQVSRAIA ADPALAARYTWANRLIAVVSDGSAVLGLGDIGPAAALPVMEGKAALFQAYGGLNAIPIVLDTKDPDEIVE TLVRLRPTFGAVNLEDISAPRCFEIERRVIEALDCPVMHDDQHGTAIVVLAALMGASKLLGRDMTSLRAV VSGAGAAGVACTNLLLAMGVLDITVLDSRGILHTSRGDMNSVKLDLARRSNPRDLTGGMAEALDGADVFL GVSGGLIPEELIATMAPGGIVFALSNPDPEIHPDIAAKYATVVATGRSDYPNQINNVLAFPGVFRGALDA GARRITQRMMVAAAEAIFSVVSDDLAVDKIVPSPLDPRVGEAVAAAVAIASDV*
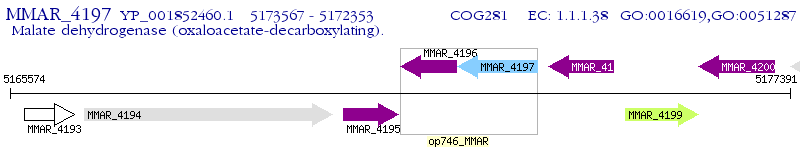
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4197 | mez_1 | - | 100% (404) | [NAD] dependent malate oxidoreductase Mez_1 [Mycobacterium marinum M] |
| M. marinum M | MMAR_3643 | mez | 2e-20 | 28.09% (388) | [NAD] dependent malate oxidoreductase Mez [Mycobacterium |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2360 | mez | 5e-21 | 28.35% (381) | malate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2214 | - | 1e-147 | 82.22% (315) | malate dehydrogenase |
| M. tuberculosis H37Rv | Rv2332 | mez | 1e-20 | 28.08% (381) | malate dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1384 | - | 1e-171 | 80.48% (374) | malate dehydrogenase |
| M. avium 104 | MAV_1381 | - | 0.0 | 83.72% (393) | NAD-dependent malic enzyme |
| M. smegmatis MC2 155 | MSMEG_5055 | - | 1e-173 | 81.58% (380) | NAD-dependent malic enzyme |
| M. thermoresistible (build 8) | TH_2059 | - | 1e-164 | 79.18% (365) | NAD-dependent malic enzyme |
| M. ulcerans Agy99 | MUL_4503 | mez_1 | 0.0 | 98.51% (404) | [NAD] dependent malate oxidoreductase Mez_1 [Mycobacterium |
| M. vanbaalenii PYR-1 | Mvan_4481 | - | 1e-178 | 82.46% (382) | malate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4197|M.marinum_M ------------------MPDTATSALKPAKALSSPSSLSPQVVIDNDEI
MUL_4503|M.ulcerans_Agy99 ------------------MPDTATSALKPAKALSSPSSLSPQVVIDNDEI
MAV_1381|M.avium_104 ----------------------------MSELLESIQSRAEQGAITDAEI
MSMEG_5055|M.smegmatis_MC2_155 ------------------------------------------MVIEDSEI
TH_2059|M.thermoresistible__bu -------------------------------------VRTPQVVVDDAEI
Mflv_2214|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4481|M.vanbaalenii_PYR-1 -------------------------MGSVTLTPVVETVRTPQVVIVDAEI
MAB_1384|M.abscessus_ATCC_1997 -------------------------------------MTASDVVVTDEEI
Mb2360|M.bovis_AF2122/97 MSDARVPRIPAALSAPSLNRGVGFTHAQRRRLGLTGRLPSAVLTLDQQAE
Rv2332|M.tuberculosis_H37Rv MSDARVPRIPAALSAPSLNRGVGFTHAQRRRLGLTGRLPSAVLTLDQQAE
MMAR_4197|M.marinum_M FRAHVDGKLSVALKAPLDTQR-------------------ALSIAYTPGV
MUL_4503|M.ulcerans_Agy99 FRAHVDGKLSVALKAPLDTQR-------------------PLSIAYTPGV
MAV_1381|M.avium_104 FAAHVGGKLSVSLTSPLDTQR-------------------ALSIAYTPGV
MSMEG_5055|M.smegmatis_MC2_155 FAAHEGGKLSVELKSPLDTQR-------------------ALSIAYTPGV
TH_2059|M.thermoresistible__bu FAHHEGGKLSVALKEPLDTQR-------------------ALSIAYTPGV
Mflv_2214|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4481|M.vanbaalenii_PYR-1 FEAHEGGKLSVALNSPLDTQR-------------------ALSIAYTPGV
MAB_1384|M.abscessus_ATCC_1997 FEAHIDGKLSVELKSPLDTQR-------------------ALSIAYTPGV
Mb2360|M.bovis_AF2122/97 RVWHQLQSLATDLGRNLLLEQLHYRHEVLYFKVLADHLPELMPVVYTPTV
Rv2332|M.tuberculosis_H37Rv RVWHQLQSLATELGRNLLLEQLHYRHEVLYFKVLADHLPELMPVVYTPTV
MMAR_4197|M.marinum_M AQVSRAIAADPALAARYTWANR-------------------LIAVVSDGS
MUL_4503|M.ulcerans_Agy99 AQVSRAIAADPALAARYTWANR-------------------LIAVVSDGS
MAV_1381|M.avium_104 AQVSRAIAADHRQAAHYTWANR-------------------LVAVVSDGS
MSMEG_5055|M.smegmatis_MC2_155 AQVSRAIAADATLAKKYTWANR-------------------LVAVVSDGS
TH_2059|M.thermoresistible__bu AQVSRAIATDRTLAAKYTWSNR-------------------LVAVVSDGS
Mflv_2214|M.gilvum_PYR-GCK --------------------------------------------MVSDGS
Mvan_4481|M.vanbaalenii_PYR-1 AQVSRAIAADKTLAARYTWAHR-------------------LVAVVSDGS
MAB_1384|M.abscessus_ATCC_1997 AQVSRAIAADASLAKKYTWAHR-------------------LVAVVSDGS
Mb2360|M.bovis_AF2122/97 GEAIQRFSDEYRGQRGLFLSIDEPDEIEEAFNTLGLGPEDVDLIVCTDAE
Rv2332|M.tuberculosis_H37Rv GEAIQRFSDEYRGQRGLFLSIDEPDEIEEAFNTLGLGPEDVDLIVCTDAE
: :*..
MMAR_4197|M.marinum_M AVLGLGDIGPAAALPVMEGKAALFQAYGGLN---AIPIVLDT-KDPDEIV
MUL_4503|M.ulcerans_Agy99 AVLGLGDIGPAAALPVMEGKAALFQAYGGLN---AIPIVLDT-KDPDEIV
MAV_1381|M.avium_104 AVLGLGDIGPAASLPVMEGKCALFQAYGGLN---AIPIVLDT-KDPDEIV
MSMEG_5055|M.smegmatis_MC2_155 AVLGLGDIGAAASLPVMEGKAALFKTFGGLD---SIPLVLDT-NDPDEIV
TH_2059|M.thermoresistible__bu AVLGLGDIGPEASLPVMEGKCALFKAFAGLD---SIPIVLDT-KDPDEIV
Mflv_2214|M.gilvum_PYR-GCK AVLGLGDIGAAASLPVMEGKSELFKTFGGLD---SIPIVLDT-KDPDEIV
Mvan_4481|M.vanbaalenii_PYR-1 AVLGLGDIGAAASLPVMEGKSALFKTFGGLD---SIPIVLDT-NDPDEIV
MAB_1384|M.abscessus_ATCC_1997 AVLGLGDIGPSASLPVMEGKSALFKTFAGLN---SIPIVLDT-KDPDEIV
Mb2360|M.bovis_AF2122/97 AILGIGDWG-VGGIQIAVGKLALYTAGGGVDPRRCLAVSLDVGTDNEQLL
Rv2332|M.tuberculosis_H37Rv AILGIGDWG-VGGIQIAVGKLALYTAGGGVDPRRCLAVSLDVGTDNEQLL
*:**:** * ..: : ** *: : .*:: .:.: **. .* ::::
MMAR_4197|M.marinum_M ETLVRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
MUL_4503|M.ulcerans_Agy99 ETLVRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
MAV_1381|M.avium_104 ETLVRLRPTFGAVN--LEDISAPRCFEIERRLIE----------------
MSMEG_5055|M.smegmatis_MC2_155 ETLVRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
TH_2059|M.thermoresistible__bu ETLIRLRPTFGAVN--LEDISAPRCFEIERRVVE----------------
Mflv_2214|M.gilvum_PYR-GCK ETLIRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
Mvan_4481|M.vanbaalenii_PYR-1 ETLIRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
MAB_1384|M.abscessus_ATCC_1997 ETLIRLRPTFGAVN--LEDISAPRCFEIERRVIE----------------
Mb2360|M.bovis_AF2122/97 ADPFYLGNRHARRRGREYDEFVSRYIETAQRLFPRAILHFEDFGPANARK
Rv2332|M.tuberculosis_H37Rv ADPFYLGNRHARRRGREYDEFVSRYIETAQRLFPRAILHFEDFGPANARK
. * .. . * ..* :* :*:.
MMAR_4197|M.marinum_M -----ALDCPVMHDDQHGTAIVVLAALMGASKLLGRDMTSLRAVVSGAGA
MUL_4503|M.ulcerans_Agy99 -----ALDCPVMHDDQHGTAIVVLAALMGASKLLGRDMTSLRAVVSGAGA
MAV_1381|M.avium_104 -----ALDCPVMHDDQHGTAIVVLAALMGAGKLLGRDMSSLKVVVSGAGA
MSMEG_5055|M.smegmatis_MC2_155 -----ALDCPVMHDDQHGTAIVVLAALLGAVKVLDRDLASLKVVVSGAGA
TH_2059|M.thermoresistible__bu -----ALDCPVMHDDQHGTAIVVLAALIGAAEVLDRDIYSLRVVISGAGA
Mflv_2214|M.gilvum_PYR-GCK -----ALDCPVMHDDQHGTAIVVLAALHGAAKVLERDMHALRVVISGAGA
Mvan_4481|M.vanbaalenii_PYR-1 -----ALDCPVMHDDQHGTAIVVLAALHGAAKALGRDMHSLRVVISGAGA
MAB_1384|M.abscessus_ATCC_1997 -----ALDCPVMHDDQHGTAIVVLAALMGASRVLGRDQQSLKVVISGAGA
Mb2360|M.bovis_AF2122/97 ILDTYGTDYCVFNDDMQGTGAVVLAAVYSGLKVTGIPLRDQTIVVFGAGT
Rv2332|M.tuberculosis_H37Rv ILDTYGTDYCVFNDDMQGTGAVVLAAVYSGLKVTGIPLRDQTIVVFGAGT
. * *::** :**. *****: .. . *: ***:
MMAR_4197|M.marinum_M AGVACTNLLL----------AMGVLDITVLDSRGILHTSRGDMNSVKLDL
MUL_4503|M.ulcerans_Agy99 AGVACTNLLL----------AMGVFDITVLDSRGTLHTSRGDMNSVKLDL
MAV_1381|M.avium_104 AGVACTNLLM----------AMGVSDITVLDSRGILHAGRDDMNDVKAAL
MSMEG_5055|M.smegmatis_MC2_155 AGVACTNILR----------NSGITDIVVLDSKGIVSGGRTDLNSFKAEL
TH_2059|M.thermoresistible__bu AGVACANILQ----------AKGIRDIVVLDSKGILHPGREDMNEVKTEL
Mflv_2214|M.gilvum_PYR-GCK AGVACANILL----------AKGITDITVLDSKGIIHPDRGDLNSFKAEL
Mvan_4481|M.vanbaalenii_PYR-1 AGVACANILL----------AKGIPDVTVLDSKGIVHTGRSDLNPFKAEL
MAB_1384|M.abscessus_ATCC_1997 AGVACANILL----------ADGINDVTVLDSKGIVHTSRDDLNEFKADL
Mb2360|M.bovis_AF2122/97 AGMGIADQIRDAMVADGATLEQAVSQIWPLDRPGLLFDDMDDLRDFQVPY
Rv2332|M.tuberculosis_H37Rv AGMGIADQIRDAMVADGATLEQAVSQIWPIDRPGLLFDDMDDLRDFQVPY
**:. :: : .: :: :* * : . *:. .:
MMAR_4197|M.marinum_M ARRSNPRDLTGG------MAEALDGADVFLGVS--GGLIPEELIATMAPG
MUL_4503|M.ulcerans_Agy99 ARRSNPRGLTGG------MAEALDGADVFLGVS--SGLIPEELIATMTPG
MAV_1381|M.avium_104 ARRTNPAGLTGS------LAEAIRGADVFLGLS--GGVVPEEMIAAMAPG
MSMEG_5055|M.smegmatis_MC2_155 AQRTNPRGLTGG------LAEALAGADVFLGVS--AGLVPEELIATMAPN
TH_2059|M.thermoresistible__bu AQRTNPRGLTAG------LAEALADADVFLGVS--AGLVPEELIASMAPK
Mflv_2214|M.gilvum_PYR-GCK AQRTNPAGRTGG------VAEALDGADMFLGLS--AGLVPEEIIATMAPG
Mvan_4481|M.vanbaalenii_PYR-1 AERTNPAGRTGG------IAEALDGADMFLGVS--AGLVPEEIIATMAPG
MAB_1384|M.abscessus_ATCC_1997 AARTNPRGLTGG------QAEALAGADVFLGVS--AGKVDESLIATMADD
Mb2360|M.bovis_AF2122/97 AKNRHQLGVAVGDRVGLSDAIKIASPTILLGCSTVYGAFTKEVVEAMTAS
Rv2332|M.tuberculosis_H37Rv AKNRHQLGVAVGDRVGLSDAIKIASPTILLGCSTVYGAFTKEVVEAMTAS
* . : . : . * : .. ::** * * . :.:: :*:
MMAR_4197|M.marinum_M G---IVFALSNPDPEIHPDIAAKYAT-----VVATGR----------SDY
MUL_4503|M.ulcerans_Agy99 G---IVFALSNPDPEIHPDIAAKYAT-----VVATGR----------SDY
MAV_1381|M.avium_104 G---IVFALSNPDPEIHPQIAAKYAT-----VVATGR----------SDF
MSMEG_5055|M.smegmatis_MC2_155 G---IVFALSNPDPEIHPDAARKYAA-----VVATGR----------SDF
TH_2059|M.thermoresistible__bu C---IVFALSNPDPEIHPDVARKYAA-----VVATGR----------SDF
Mflv_2214|M.gilvum_PYR-GCK G---IVFALSNPDPEIHPDAARKYAS-----VVATGR----------SDF
Mvan_4481|M.vanbaalenii_PYR-1 G---IVFALSNPDPEIHPDAARKYAA-----VVATGR----------SDF
MAB_1384|M.abscessus_ATCC_1997 S---IVFACSNPDPEIHPHVAAKYAA-----IVATGR----------SDF
Mb2360|M.bovis_AF2122/97 CKHPMIFPLSNPTSRMEAIPADVLAWSNGRALLATGSPVAPVEFDETTYV
Rv2332|M.tuberculosis_H37Rv CKHPMIFPLSNPTSRMEAIPADVLAWSNGRALLATGSPVAPVEFDETTYV
::*. *** ..:.. * * ::*** :
MMAR_4197|M.marinum_M PNQINNVLAFPGVFRGALDAGARRITQRMMVAAAEAIFSVVSDDLAVD--
MUL_4503|M.ulcerans_Agy99 PNQINNVLAFPGVFRGALDAGARRITQRMMVAAAEAIFSVVSDDLAVD--
MAV_1381|M.avium_104 PNQINNVLAFPGVFRGALEAGARRITEKMMVAAAEAIFSVVSDDLAPD--
MSMEG_5055|M.smegmatis_MC2_155 PNQINNVLAFPGVFRGALDAGARRITEEMKVAAAHAIFSVVGDDLGVD--
TH_2059|M.thermoresistible__bu PNQINNVLAFPGVFRGALDAGARRITEEMKVAAARAIFSVIGDDLGVDXX
Mflv_2214|M.gilvum_PYR-GCK PNQINNVLAFPGVFRGALDAGARRITEKMKVAAAEAIFSVVGDDLSVD--
Mvan_4481|M.vanbaalenii_PYR-1 PNQINNVLAFPGVFRGALDAGARRITEKMKVAAAEAIFSVVGDDLAVD--
MAB_1384|M.abscessus_ATCC_1997 PNQINNVLAFPGVFRGALDAEATKITERMKVAAAEAIFSVVEPELAPN--
Mb2360|M.bovis_AF2122/97 IGQANNVLAFPGIGLGVIVAGARLITRRMLHAAAKAIAHQANPTNPGD--
Rv2332|M.tuberculosis_H37Rv IGQANNVLAFPGIGLGVIVAGARLITRRMLHAAAKAIAHQANPTNPGD--
.* ********: *.: * * **..* ***.** :
MMAR_4197|M.marinum_M --------------------------------------------------
MUL_4503|M.ulcerans_Agy99 --------------------------------------------------
MAV_1381|M.avium_104 --------------------------------------------------
MSMEG_5055|M.smegmatis_MC2_155 --------------------------------------------------
TH_2059|M.thermoresistible__bu XASPVFGPTPVTPIRVRNRSRASVSGKPYNVIESSRTIIAVINRASPPRR
Mflv_2214|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4481|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1384|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2360|M.bovis_AF2122/97 --------------------------------------------------
Rv2332|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4197|M.marinum_M ---------------------------------------KIVPSPLDPRV
MUL_4503|M.ulcerans_Agy99 ---------------------------------------KIVPSPLDPRV
MAV_1381|M.avium_104 ---------------------------------------RIVPSPLDTRV
MSMEG_5055|M.smegmatis_MC2_155 ---------------------------------------HIVPSALDPRV
TH_2059|M.thermoresistible__bu SVDRVAGVAITLYPTPAASITAWSSAISSTSPRTEAITGHPATSAAAPPS
Mflv_2214|M.gilvum_PYR-GCK ---------------------------------------HIVPSALDPRV
Mvan_4481|M.vanbaalenii_PYR-1 ---------------------------------------HIVPSALDPRV
MAB_1384|M.abscessus_ATCC_1997 ---------------------------------------KIVPSPLDPRV
Mb2360|M.bovis_AF2122/97 ---------------------------------------SLLPDVQNLRA
Rv2332|M.tuberculosis_H37Rv ---------------------------------------SLLPDVQNLRA
..
MMAR_4197|M.marinum_M G----------------EAVAAAVAIASDV--------------------
MUL_4503|M.ulcerans_Agy99 G----------------EAVAAAVAIASDV--------------------
MAV_1381|M.avium_104 G----------------DAVATAVALATEVSDAPAG--------------
MSMEG_5055|M.smegmatis_MC2_155 G----------------PAVAAAVAEASGV--------------------
TH_2059|M.thermoresistible__bu GGRGTCVAVGRPGPRPWPSARASAPASSGRSPTPTRPRHQPVSVLSVAQG
Mflv_2214|M.gilvum_PYR-GCK G----------------PAVAAAVAAASED--------------------
Mvan_4481|M.vanbaalenii_PYR-1 G----------------PAVAAAVAAASEE--------------------
MAB_1384|M.abscessus_ATCC_1997 G----------------PAVAAAVQAVAHESD------------------
Mb2360|M.bovis_AF2122/97 IS---------------TTVAEAVYRAAVQDGVASRTHDDVRQAIVDTMW
Rv2332|M.tuberculosis_H37Rv IS---------------TTVAEAVYRAAVQDGVASRTHDDVRQAIVDTMW
:. :. :
MMAR_4197|M.marinum_M ------
MUL_4503|M.ulcerans_Agy99 ------
MAV_1381|M.avium_104 ------
MSMEG_5055|M.smegmatis_MC2_155 ------
TH_2059|M.thermoresistible__bu FSSPSR
Mflv_2214|M.gilvum_PYR-GCK ------
Mvan_4481|M.vanbaalenii_PYR-1 ------
MAB_1384|M.abscessus_ATCC_1997 ------
Mb2360|M.bovis_AF2122/97 LPAYD-
Rv2332|M.tuberculosis_H37Rv LPAYD-