For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSITDIGGIRVGHYQRLDPDASLGAGWASGVTVVLAPPGTVGAVDCRGGAPGTRETDLLDPVNSVRFVDA VLLAGGSAYGLAAADGVMRWLEEHQRGVAMGAGVVPIVPGAVIFDLPVGGWGQRPTAEFGYLACDAAVSD QDPGATASLAVGTVGAGVGARAGVLKGGVGTASVKLPSGVTVGAVVVVNSVGNVVDPATGLPWMAELSGE FGLTRPPADQLDELARLPSPLDPQNAPQDRPLNTTIAVVATDAALSPAGCQRVAIAAQDGLARTIRPAHT PLDGDTVFALATGAVQVPPDPAIPAALSPETGLLTAVGAAAADCLAQAVLVGVIAAESVAGIPSYRDVLP GALQTSGTESR*
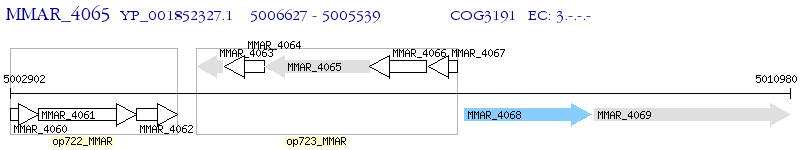
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4065 | dmpA | - | 100% (362) | L-aminopeptidase/D-esterase DmpA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1368 | - | 1e-155 | 77.90% (353) | hydrolase |
| M. gilvum PYR-GCK | Mflv_2351 | - | 1e-133 | 69.52% (351) | peptidase S58, DmpA |
| M. tuberculosis H37Rv | Rv1333 | - | 1e-155 | 77.90% (353) | hydrolase |
| M. leprae Br4923 | MLBr_01167 | - | 1e-146 | 75.07% (357) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_1477 | - | 1e-113 | 61.82% (351) | hypothetical protein MAB_1477 |
| M. avium 104 | MAV_1553 | - | 1e-155 | 78.53% (354) | peptidase family protein T4 |
| M. smegmatis MC2 155 | MSMEG_4908 | - | 1e-142 | 73.52% (355) | endo-type 6-aminohexanoate oligomer hydrolase |
| M. thermoresistible (build 8) | TH_1833 | - | 8e-10 | 25.41% (362) | D-aminopeptidase |
| M. ulcerans Agy99 | MUL_3929 | dmpA | 0.0 | 99.45% (362) | L-aminopeptidase/D-esterase DmpA |
| M. vanbaalenii PYR-1 | Mvan_4294 | - | 1e-135 | 70.82% (353) | peptidase S58, DmpA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2351|M.gilvum_PYR-GCK -----------------MSGSITDVGGIRVGHHHAIDADSTLGSG---WA
Mvan_4294|M.vanbaalenii_PYR-1 -----------------MSGSITDVGGIRVGHHHRLDDDAGMGSG---WA
MSMEG_4908|M.smegmatis_MC2_155 ------------------MGSITDVAGILVGHHHRLDPDAVLGSG---WA
Mb1368|M.bovis_AF2122/97 ------------------MNSITDVGGIRVGHYQRLDPDASLGAG---WA
Rv1333|M.tuberculosis_H37Rv ------------------MNSITDVGGIRVGHYQRLDPDASLGAG---WA
MMAR_4065|M.marinum_M ------------------MSSITDIGGIRVGHYQRLDPDASLGAG---WA
MUL_3929|M.ulcerans_Agy99 ------------------MSSITDIGGIRVGHYQRLDPDASLGAG---WA
MAV_1553|M.avium_104 ------------------MNAITDVGGIRVGHHQRLDPDASLGAG---WA
MLBr_01167|M.leprae_Br4923 -----------MILMAAPMNSITDISGIQVGHYHRLDPDASLGAG---WA
MAB_1477|M.abscessus_ATCC_1997 --------------MSLFSGAITDVPGILVGHHQRVDADASLGAG---WA
TH_1833|M.thermoresistible__bu VARARDYGATIGVLPTGPNNAITDVAGVRVGHTTLIEGSGPRVTGQGPVR
.:***: *: *** :: .. :*
Mflv_2351|M.gilvum_PYR-GCK TGTTVVLTPPGTVGAVDSRGGAPGSRETDLLAPANSVRHVDAVVLTGGS-
Mvan_4294|M.vanbaalenii_PYR-1 TGTTVVLTPPGTVGAVDGRGGAPGTRETDLLDPSNSVRHVDAVVLTGGS-
MSMEG_4908|M.smegmatis_MC2_155 SGTTVVLTPPGTVGAVDGRGGAPGTRETDLLDPCNTVRHVDAVVLTGGS-
Mb1368|M.bovis_AF2122/97 CGVTVVLPPPGTVGAVDCRGGAPGTRETDLLDPANSVRFVDALLLAGGS-
Rv1333|M.tuberculosis_H37Rv CGVTVVLPPPGTVGAVDCRGGAPGTRETDLLDPANSVRFVDALLLAGGS-
MMAR_4065|M.marinum_M SGVTVVLAPPGTVGAVDCRGGAPGTRETDLLDPVNSVRFVDAVLLAGGS-
MUL_3929|M.ulcerans_Agy99 SGVTVVLAPPGTVGAVDCRGGAPGTRETDLLDPVNSVRFVDAVLLAGGS-
MAV_1553|M.avium_104 CGVTVVLTPPGTVGAVDCRGGAPGTRETDLLDPANTVRFVDAVLLAGGS-
MLBr_01167|M.leprae_Br4923 CGVTVVLTPPGTVGAVDCRGGVPGTRETDLLDPANSVRFVDAVLLAGGS-
MAB_1477|M.abscessus_ATCC_1997 TGTTVVLAPPGTTGAVDVRGGAPGTRESDLLDPANTVATVDAVVLTGGS-
TH_1833|M.thermoresistible__bu TGVTVVIPRDEPWNHPLFAAAHRLNGNGELTGQHWVEESGQLTSYIGLTN
*.***:. . . .. . : :* : * :
Mflv_2351|M.gilvum_PYR-GCK AFGLAAADGVMAWLEEQGRGVALDGGVVPIVPAAVIFDLP-VGGWRCRPD
Mvan_4294|M.vanbaalenii_PYR-1 AFGLAAADGVMTWLEEQNRGVALDGGVVPIVPAAVIFDLP-VGGWKCRPD
MSMEG_4908|M.smegmatis_MC2_155 AYGLAAADGVMTWLEEQGRGVAMEGGVVPIVPAAVIFDLP-VGAWGARPT
Mb1368|M.bovis_AF2122/97 AYGLAAADGVMRWLEEHRRGVAMDSGVVPIVPGAVIFDLP-VGGWNCRPT
Rv1333|M.tuberculosis_H37Rv AYGLAAADGVMRWLEEHRRGVAMDSGVVPIVPGAVIFDLP-VGGWNCRPT
MMAR_4065|M.marinum_M AYGLAAADGVMRWLEEHQRGVAMGAGVVPIVPGAVIFDLP-VGGWGQRPT
MUL_3929|M.ulcerans_Agy99 AYGLAAADGVMRWLEEHQRGVAMGTGVVPIVPGAVIFDLP-VGGWGQRPT
MAV_1553|M.avium_104 AYGLAAADGVMRWLEEHDRGVAMDGGVVPIVPGAVIFDLP-VGGWQCRPT
MLBr_01167|M.leprae_Br4923 AYGLAAADGVMRWLEEHERGVVMLGGVVPIVPGAVIFDLS-VGDFHCRPT
MAB_1477|M.abscessus_ATCC_1997 AYGLAAADGVMRWLEERGRGVPLSGGTVPIVPAAVIFDLP-VGAWKNRPD
TH_1833|M.thermoresistible__bu THSVGVVRDALIAHEKNARGADEFFFALPVVGETCDGILNDINGFHVKPE
:..:... ..: *:. **. .:*:* : * :. : :*
Mflv_2351|M.gilvum_PYR-GCK AAFGYRAAETAGAD--------AAIGSVGAGVGARAGVLKGGVGTASTTL
Mvan_4294|M.vanbaalenii_PYR-1 ADFGYRAAAAAGTD--------AGIGTVGAGVGARAGVLKGGVGTASVTL
MSMEG_4908|M.smegmatis_MC2_155 AQFGYEAAASAGTE--------FALGTVGAGVGARVGVLKGGVGTASMTL
Mb1368|M.bovis_AF2122/97 ADFGYSACAAAGVD--------VAVGTVGVGVGARAGALKGGVGTASATL
Rv1333|M.tuberculosis_H37Rv ADFGYSACAAAGVD--------VAVGTVGVGVGARAGALKGGVGTASATL
MMAR_4065|M.marinum_M AEFGYLACDAAVSDQDPGATASLAVGTVGAGVGARAGVLKGGVGTASVKL
MUL_3929|M.ulcerans_Agy99 AEFGYLACDAAVSDQDPGATASLAVGTVGAGVGARAGVLKGGVGTASVKL
MAV_1553|M.avium_104 AEFGYLACAAARDDDAP------AVGTVGAGVGARAGALKGGVGTASRSL
MLBr_01167|M.leprae_Br4923 AEFGYLACQAAYDAAVGGQDATVAVGTVGAGVGARAGVLKGGVGTASITL
MAB_1477|M.abscessus_ATCC_1997 TEFGYRAAELADDAP--------EWGAVGAGAGARAGTLKGGVGSASVNV
TH_1833|M.thermoresistible__bu HVAAAIDAAADGPVP---------EGAVGGGTGMIAHGYKGGIGTSSRVV
. . *:** *.* . ***:*::* :
Mflv_2351|M.gilvum_PYR-GCK DSGVTVGALVVVNSAGDVVDPRTGLPWLAAHAEEFGLVAPPADQRSAYSD
Mvan_4294|M.vanbaalenii_PYR-1 ESGVTVGALVVVNSAGDVIDPGSGLPWQASHIEEFGLVRPPADELAAYAD
MSMEG_4908|M.smegmatis_MC2_155 ENGVTVGAVVVVNAAGDAVDPATGLPWMAEYVEEFGLIPPPADRLTGYAD
Mb1368|M.bovis_AF2122/97 QSGVTVGVLAVVNAAGNVVDPATGLPWMADLVGEFALRAPPAEQIAALAQ
Rv1333|M.tuberculosis_H37Rv QSGVTVGVLAVVNAAGNVVDPATGLPWMADLVGEFALRAPPAEQIAALAQ
MMAR_4065|M.marinum_M PSGVTVGAVVVVNSVGNVVDPATGLPWMAELSGEFGLTRPPADQLDELAR
MUL_3929|M.ulcerans_Agy99 PSGVTVGAVVVVNSVGNVVDPATGLPWMAELSGEFGLTRPPADQLDELAR
MAV_1553|M.avium_104 PSGVTVGALAVVNSAGEVVDRATGLPWMTDLVEQFGLRPPPVEQIEAFAA
MLBr_01167|M.leprae_Br4923 ESGPTVGAVVVVNSVGDVVDRATGLPWMTDLIDEFALRPPSPEQIAGFAQ
MAB_1477|M.abscessus_ATCC_1997 GDGIVVGALVVANPVGLVINPATGRPWTADILPELS--APPAAEIEKLRD
TH_1833|M.thermoresistible__bu G-DYTVGVLVQANHGARERLTIDGVP--------VGRLLPEPPSPAAMPG
. .**.:. .* . * * .. *
Mflv_2351|M.gilvum_PYR-GCK RHGELS--------PLNTTIAVVATDAALSKAACSRVAVAAQDGLARTIN
Mvan_4294|M.vanbaalenii_PYR-1 RHGELG--------LLNTTIAVVATDAALSKAACRRVAVAAQDGLARTIN
MSMEG_4908|M.smegmatis_MC2_155 LRTELS--------PLNTTIAVVATDAELSPAACKRVAVASHDGLARTLR
Mb1368|M.bovis_AF2122/97 LSSPLG----AFNTPFNTTIGVIACDAALSPAACRRIAIAAHDGLARTIR
Rv1333|M.tuberculosis_H37Rv LSSPLG----AFNTPFNTTIGVIACDAALSPAACRRIAIAAHDGLARTIR
MMAR_4065|M.marinum_M LPSPLDPQNAPQDRPLNTTIAVVATDAALSPAGCQRVAIAAQDGLARTIR
MUL_3929|M.ulcerans_Agy99 LPSPLDPQDAPQDRPLNTTIAVVATDAALSPAGCQRVAIAAQDGLARTIR
MAV_1553|M.avium_104 LPSPSN----PLDNPLNTVIAVVATDAALSAAACRRVAVAAHDGLARSIR
MLBr_01167|M.leprae_Br4923 LKSPLS--------ALNTTIGVVATDATLSPAACQRVAMAAQDGLARTIR
MAB_1477|M.abscessus_ATCC_1997 LPDKSV--------SLNTTIGVVATNAALSKAQCRRVAIAAHDGLARAIR
TH_1833|M.thermoresistible__bu RPPEGT----------GSIIVLVATDAPLIPTQCARLAQRSALGIARTGG
.: * ::* :* * : * *:* : *:**:
Mflv_2351|M.gilvum_PYR-GCK PCHTPQDGDTVFALATG-----AVEVNPDPTTPASMTPELPLVTAVGAAA
Mvan_4294|M.vanbaalenii_PYR-1 PCHTPLDGDTVFALATG-----AVEVNPDPTTPASMTPELPLVTAVGAAA
MSMEG_4908|M.smegmatis_MC2_155 PCHTPLDGDTVFALATG-----RVAVPPDPKTPTAMSPETRLVTQVGAAA
Mb1368|M.bovis_AF2122/97 PAHTPLDGDTVFALATG-----AVAVPPEAGVPAALSPETQLVTAVGAAA
Rv1333|M.tuberculosis_H37Rv PAHTPLDGDTVFALATG-----AVAVPPEAGVPAALSPETQLVTAVGAAA
MMAR_4065|M.marinum_M PAHTPLDGDTVFALATG-----AVQVPPDPAIPAALSPETGLLTAVGAAA
MUL_3929|M.ulcerans_Agy99 PAHTPLDGDTVFALATG-----AVQVPPDPAIPAALSPETGLLTAVGAAA
MAV_1553|M.avium_104 PAHTPVDGDTVFVLATG-----AVEVPPEADTPAAFSPETRLATEVGAAA
MLBr_01167|M.leprae_Br4923 PAHTALDGDTVFALATG-----AVEATATADVPVAMSPETGLITEVGAAA
MAB_1477|M.abscessus_ATCC_1997 PAHTPMDGDTLYVLATG-----TGEGDADP-----------LTAAVGIAA
TH_1833|M.thermoresistible__bu AGENG-SGDMAFCFTTGNTFAGDFAATAGPDVVEVRMLANRLIDRLFYAA
. .. .** : ::** . . * : **
Mflv_2351|M.gilvum_PYR-GCK AEVLARAVLVAVLAAERVAG-IPTYRELLPGAFA-----------
Mvan_4294|M.vanbaalenii_PYR-1 AEVLARAVLVGVMAADRVAG-IPTYRGLLPGAFE-----------
MSMEG_4908|M.smegmatis_MC2_155 ADCLARAVLVGVLAAESVAG-IPTYRDMLPGAFE-----------
Mb1368|M.bovis_AF2122/97 ADCLARAVLAGVLNAQPVAG-IPTYRDMFPGAFGS----------
Rv1333|M.tuberculosis_H37Rv ADCLARAVLAGVLNAQPVAG-IPTYRDMFPGAFGS----------
MMAR_4065|M.marinum_M ADCLAQAVLVGVIAAESVAG-IPSYRDVLPGALQTSGTESR----
MUL_3929|M.ulcerans_Agy99 ADCLAQAVLVGVIAAESVAG-IPSYRDVLPGALQTSGTESR----
MAV_1553|M.avium_104 ADCLAHAVLGGVLAADSVAG-IPSYRDVLPGALGR----------
MLBr_01167|M.leprae_Br4923 DDCLARAVLVAVLAAESVAG-IPTYCGMFPGAFGTTIGGGNR---
MAB_1477|M.abscessus_ATCC_1997 ADCVERAVVRAVVGAASVAG-IPAYRDVLPGAFS-----------
TH_1833|M.thermoresistible__bu IEATEEAIVNALFAAETMEGPEGLVVTALPREQTAELLRRYRQLR
: .*:: .:. * : * :*